- "markdown": "---\ntitle: \"Exploring the Wide World of ggplot2 Extensions\"\nauthor: \n - \"Eric R. Scott\"\n - \"Kristina Riemer\"\n - \"Renata Diaz\"\ndate: 2024-06-20\nformat: \n uaz-revealjs: default\nexecute: \n echo: true\neditor: visual\n---\n\n\n## Learning Objectives\n\n- Understand where to find packages that extend `ggplot2`\n- Assemble multi-panel figures with `patchwork`\n- Make animated plots with `gganimate`\n- Visualize distributions with `ggdist`\n- Plot network data with `ggraph`\n\n## Packages\n\n\n::: {.cell}\n\n```{.r .cell-code}\nlibrary(ggplot2)\nlibrary(palmerpenguins) #for example dataset\nlibrary(patchwork) #for multi-panel figures\nlibrary(gridGraphics) #for combining ggplot2 and base R figures\nlibrary(gganimate) #for animated plots\n```\n:::\n\n\n# Extensions to `ggplot2`\n\nThere are many kinds of extensions to `ggplot2`, ranging from simple to complex, from familiar to transformative.\n\n## General-use and modular\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code code-line-numbers=\"|1,4|2,5\"}\nlibrary(ggbeeswarm)\nlibrary(ggthemes)\nggplot(penguins, aes(x = species, y = body_mass_g)) +\n geom_beeswarm() +\n theme_economist()\n```\n\n::: {.cell-output-display}\n{fig-align='center' width=960}\n:::\n:::\n\n\n::: notes\n`geom_beeswarm()` can easily be combined with other `geom_`s\n:::\n\n## \"All-in-one\" functions\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code code-line-numbers=\"|1,2|3\"}\nlibrary(ggstatsplot)\nggbetweenstats(penguins, species, body_mass_g) +\n scale_y_continuous(\"Body Mass (g)\", n.breaks = 10)\n```\n\n::: {.cell-output-display}\n{fig-align='center' width=960}\n:::\n:::\n\n\n::: notes\nWith great power comes great responsibility—make sure you trust these stats!\\\nNotice that the output is a ggplot object, so you can continue to add layers to it.\n:::\n\n## Transformative & field-specific\n\n::: {layout-ncol=\"2\"}\n[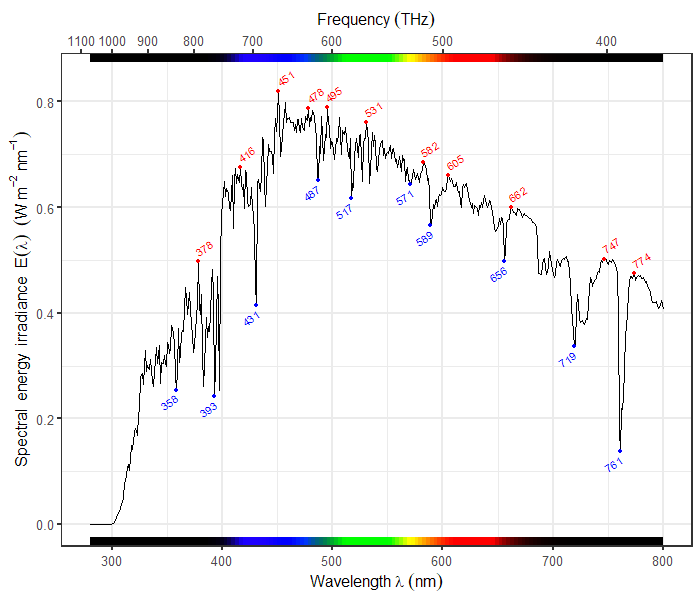{width=\"100%\"}](https://docs.r4photobiology.info/ggspectra)\n\n[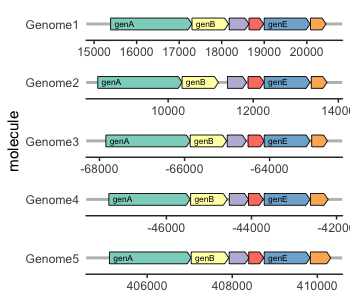{width=\"100%\"}](https://wilkox.org/gggenes)\n:::\n\n## Finding Extensions\n\n- Browse the `ggplot2` [extensions gallery](https://exts.ggplot2.tidyverse.org/gallery/)\n\n- Check out the [Awesome `ggplot2`](https://github.com/erikgahner/awesome-ggplot2#readme) list\n\n- [The R Graph Gallery](https://r-graph-gallery.com/)\n\n- Google search with \"ggplot2\" keyword\n\n# Package Demos\n\n## `patchwork`\n\n`patchwork` allows you to compose multi-panel figures with ease\n\n\n::: {.cell layout-align=\"center\"}\n::: {.cell-output-display}\n{fig-align='center' width=960}\n:::\n:::\n\n\n::: notes\nThings to notice:\n\n- Plots areas are aligned despite one not having a y-axis label\n- Only one legend\n- Labels for panels A, B, C\n:::\n\n## Example plots\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 <- \n ggplot(penguins, aes(x = body_mass_g, y = bill_length_mm, color = species)) +\n geom_point()\np2 <-\n ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, color = species)) +\n geom_point()\np3 <- \n ggplot(penguins, aes(x = body_mass_g)) + \n facet_wrap(vars(island)) +\n geom_histogram()\n```\n:::\n\n\n## Combine plots\n\n::: columns\n::: {.column width=\"50%\"}\n::: nonincremental\n- `+` wraps plots\n- `|` combines plots horizontally\n- `/` combines plots vertically\n- `()` can be used to nest operations\n:::\n:::\n\n::: {.column width=\"50%\"}\n\n::: {.cell}\n\n```{.r .cell-code}\n(p1 | p2) / p3\n```\n\n::: {.cell-output-display}\n{width=480}\n:::\n:::\n\n:::\n:::\n\n## Combine guides\n\nIf plots have *identical* guides, you can combine them with `plot_layout(guides = \"collect\")`\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 + p2 + plot_layout(guides = \"collect\")\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n::: notes\nidentical means same scale, same title, same glyph\n:::\n\n## Combining axes\n\nAs of the most recent version of `patchwork` (v1.2.0) you can also combine identical axes.\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 + p2 + plot_layout(guides = \"collect\", axes = \"collect\")\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n## Controlling layout\n\nFor more options for controlling layout, see the related vignette on the package website.\n\n- Adding empty areas\n- Inset plots\n- Adjusting widths and heights\n\n## Tags\n\nYou can add \"tags\" to each panel with `plot_annotation()`\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 + p2 + plot_annotation(tag_levels = \"A\", tag_suffix = \")\")\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n## Modifying all panels\n\nYou can use the `&` operator instead of `+` to modify **all** elements of a multi-panel figure.\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 + p2 & theme_bw() & scale_color_viridis_d()\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n## Using with base R plots\n\nYou can even combine `ggplot2` plots with base R plots with a special syntax\n\n\n::: {.cell}\n\n```{.r .cell-code}\np1 + ~hist(penguins$bill_depth_mm, main = \"\")\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n## `gganimate`\n\nDisplay transitions between states of a variable\n\nUseful for showing trends in time series & spatial data\n\n::: callout-tip\n## Advice from the experts!\n\n\"Graphic elements should only transition between instances of the same underlying phenomenon\"\n:::\n\n## Example\n\nBody size distribution of penguins changing over the years\n\n\n::: {.cell}\n\n```{.r .cell-code}\nggplot(penguins, aes(x = body_mass_g, col = species)) +\n geom_density() +\n facet_wrap(~year)\n```\n\n::: {.cell-output-display}\n{width=960}\n:::\n:::\n\n\n## Show annual transition\n\n\n::: {.cell}\n\n```{.r .cell-code}\nlibrary(gganimate)\n\nggplot(penguins, aes(x = body_mass_g, col = species)) +\n geom_density() +\n transition_time(year) +\n ggtitle('Year: {frame_time}')\n```\n\n::: {.cell-output-display}\n\n:::\n:::\n\n\n## Add history\n\n\n::: {.cell}\n\n```{.r .cell-code}\nggplot(penguins, aes(x = body_mass_g, col = species)) +\n geom_density() +\n transition_time(year) +\n ggtitle('Year: {frame_time}') +\n shadow_wake(wake = 0.3, wrap = FALSE)\n```\n\n::: {.cell-output-display}\n\n:::\n:::\n\n\n## Modify transition speed\n\n\n::: {.cell}\n\n```{.r .cell-code}\nggplot(penguins, aes(x = body_mass_g, col = species)) +\n geom_density() +\n transition_time(year) +\n ggtitle('Year: {frame_time}') +\n ease_aes(\"quintic-in-out\")\n```\n\n::: {.cell-output-display}\n\n:::\n:::\n\n\n## Have axes follow data\n\n\n::: {.cell}\n\n```{.r .cell-code}\nggplot(penguins, aes(x = body_mass_g, col = species)) +\n geom_density() +\n transition_time(year) +\n ggtitle('Year: {frame_time}') +\n view_follow()\n```\n\n::: {.cell-output-display}\n\n:::\n:::\n\n\n## Save file\n\n\n::: {.cell}\n\n```{.r .cell-code}\nanim_save(\"~/Desktop/test_anim.gif\")\n```\n:::\n\n\n## Extensions Resources\n\n`patchwork`\n\n- [Package website](https://patchwork.data-imaginist.com/index.html)\n\n`gganimate`\n\n- [Cheat sheet](https://rstudio.github.io/cheatsheets/gganimate.pdf)\n- [Website](https://gganimate.com)\n\n## Getting Help\n\n- Our [drop-in hours](https://datascience.cct.arizona.edu/drop-in-hours)\n\n- [UA Data Science Slack](https://join.slack.com/t/uadatascience/shared_invite/zt-1xjxht9k0-E489WA6axO_SRVeKQh3cXg)\n\n# Anything we missed?\n\nGot a `ggplot2` question we didn't cover in this workshop series?\nLet's figure it out together!\n",
0 commit comments