|
| 1 | +--- |
| 2 | +title: "Exploring the Wide World of ggplot2 Extensions" |
| 3 | +author: |
| 4 | + - "Eric R. Scott" |
| 5 | + - "Kristina Riemer" |
| 6 | + - "Renata Diaz" |
| 7 | +date: 2024-06-20 |
| 8 | +format: |
| 9 | + uaz-revealjs: default |
| 10 | +execute: |
| 11 | + echo: true |
| 12 | +editor: visual |
| 13 | +--- |
| 14 | + |
| 15 | +## Learning Objectives |
| 16 | + |
| 17 | +- Understand where to find packages that extend `ggplot2` |
| 18 | +- Assemble multi-panel figures with `patchwork` |
| 19 | +- Make animated plots with `gganimate` |
| 20 | +- Visualize distributions with `ggdist` |
| 21 | +- Plot network data with `ggraph` |
| 22 | + |
| 23 | +## Packages |
| 24 | + |
| 25 | +```{r} |
| 26 | +library(ggplot2) |
| 27 | +library(palmerpenguins) #for example dataset |
| 28 | +library(patchwork) #for multi-panel figures |
| 29 | +library(gridGraphics) #for combining ggplot2 and base R figures |
| 30 | +library(gganimate) #for animated plots |
| 31 | +``` |
| 32 | + |
| 33 | +# Extensions to `ggplot2` |
| 34 | + |
| 35 | +There are many kinds of extensions to `ggplot2`, ranging from simple to complex, from familiar to transformative. |
| 36 | + |
| 37 | +## General-use and modular |
| 38 | + |
| 39 | +```{r} |
| 40 | +#| code-line-numbers: "|1,4|2,5" |
| 41 | +#| fig-align: center |
| 42 | +library(ggbeeswarm) |
| 43 | +library(ggthemes) |
| 44 | +ggplot(penguins, aes(x = species, y = body_mass_g)) + |
| 45 | + geom_beeswarm() + |
| 46 | + theme_economist() |
| 47 | +
|
| 48 | +``` |
| 49 | + |
| 50 | +::: notes |
| 51 | +`geom_beeswarm()` can easily be combined with other `geom_`s |
| 52 | +::: |
| 53 | + |
| 54 | +## "All-in-one" functions |
| 55 | + |
| 56 | +```{r} |
| 57 | +#| code-line-numbers: "|1,2|3" |
| 58 | +#| fig-align: center |
| 59 | +library(ggstatsplot) |
| 60 | +ggbetweenstats(penguins, species, body_mass_g) + |
| 61 | + scale_y_continuous("Body Mass (g)", n.breaks = 10) |
| 62 | +``` |
| 63 | + |
| 64 | +::: notes |
| 65 | +With great power comes great responsibility—make sure you trust these stats!\ |
| 66 | +Notice that the output is a ggplot object, so you can continue to add layers to it. |
| 67 | +::: |
| 68 | + |
| 69 | +## Transformative & field-specific |
| 70 | + |
| 71 | +::: {layout-ncol="2"} |
| 72 | +[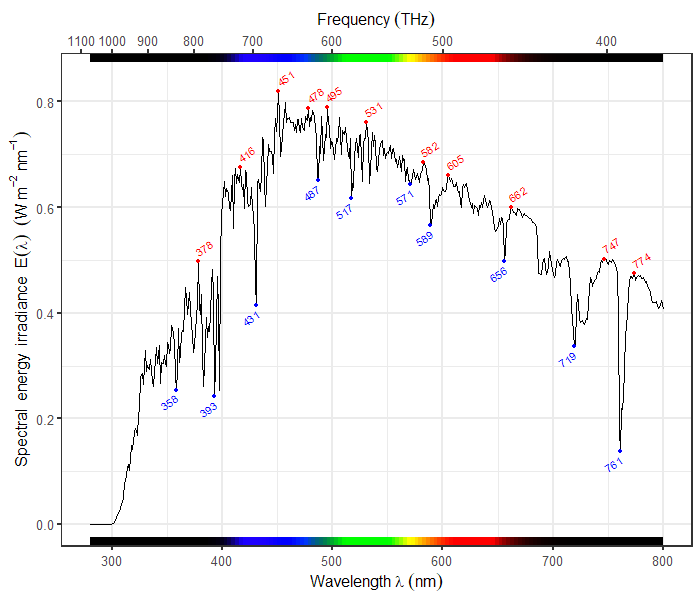{width="100%"}](https://docs.r4photobiology.info/ggspectra) |
| 73 | + |
| 74 | +[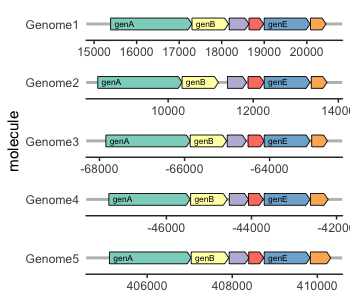{width="100%"}](https://wilkox.org/gggenes) |
| 75 | +::: |
| 76 | + |
| 77 | +## Finding Extensions |
| 78 | + |
| 79 | +- Browse the `ggplot2` [extensions gallery](https://exts.ggplot2.tidyverse.org/gallery/) |
| 80 | + |
| 81 | +- Check out the [Awesome `ggplot2`](https://github.com/erikgahner/awesome-ggplot2#readme) list |
| 82 | + |
| 83 | +- [The R Graph Gallery](https://r-graph-gallery.com/) |
| 84 | + |
| 85 | +- Google search with "ggplot2" keyword |
| 86 | + |
| 87 | +# Package Demos |
| 88 | + |
| 89 | +## `patchwork` |
| 90 | + |
| 91 | +`patchwork` allows you to compose multi-panel figures with ease |
| 92 | + |
| 93 | +```{r} |
| 94 | +#| echo: false |
| 95 | +#| fig-align: center |
| 96 | +library(patchwork) |
| 97 | +p1 <- ggplot(penguins, aes(x = body_mass_g, y = bill_length_mm, color = species)) + |
| 98 | + geom_point() |
| 99 | +p2 <- ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, color = species)) + |
| 100 | + geom_point() |
| 101 | +p3 <- ggplot(penguins, aes(x = body_mass_g)) + |
| 102 | + facet_wrap(vars(island)) + |
| 103 | + geom_histogram() + |
| 104 | + theme(axis.title.y = element_blank()) |
| 105 | +
|
| 106 | +(p1 + p2 + plot_layout(guides = "collect")) / p3 + |
| 107 | + plot_annotation(tag_levels = "A", tag_suffix = ")") |
| 108 | +``` |
| 109 | + |
| 110 | +::: notes |
| 111 | +Things to notice: |
| 112 | + |
| 113 | +- Plots areas are aligned despite one not having a y-axis label |
| 114 | +- Only one legend |
| 115 | +- Labels for panels A, B, C |
| 116 | +::: |
| 117 | + |
| 118 | +## Example plots |
| 119 | + |
| 120 | +```{r} |
| 121 | +p1 <- |
| 122 | + ggplot(penguins, aes(x = body_mass_g, y = bill_length_mm, color = species)) + |
| 123 | + geom_point() |
| 124 | +p2 <- |
| 125 | + ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, color = species)) + |
| 126 | + geom_point() |
| 127 | +p3 <- |
| 128 | + ggplot(penguins, aes(x = body_mass_g)) + |
| 129 | + facet_wrap(vars(island)) + |
| 130 | + geom_histogram() |
| 131 | +``` |
| 132 | + |
| 133 | +## Combine plots |
| 134 | + |
| 135 | +::: columns |
| 136 | +::: {.column width="50%"} |
| 137 | +::: nonincremental |
| 138 | +- `+` wraps plots |
| 139 | +- `|` combines plots horizontally |
| 140 | +- `/` combines plots vertically |
| 141 | +- `()` can be used to nest operations |
| 142 | +::: |
| 143 | +::: |
| 144 | + |
| 145 | +::: {.column width="50%"} |
| 146 | +```{r} |
| 147 | +#| fig-width: 5 |
| 148 | +#| fig-height: 5 |
| 149 | +(p1 | p2) / p3 |
| 150 | +``` |
| 151 | +::: |
| 152 | +::: |
| 153 | + |
| 154 | +## Combine guides |
| 155 | + |
| 156 | +If plots have *identical* guides, you can combine them with `plot_layout(guides = "collect")` |
| 157 | + |
| 158 | +```{r} |
| 159 | +p1 + p2 + plot_layout(guides = "collect") |
| 160 | +``` |
| 161 | + |
| 162 | +::: notes |
| 163 | +identical means same scale, same title, same glyph |
| 164 | +::: |
| 165 | + |
| 166 | +## Combining axes |
| 167 | + |
| 168 | +As of the most recent version of `patchwork` (v1.2.0) you can also combine identical axes. |
| 169 | + |
| 170 | +```{r} |
| 171 | +p1 + p2 + plot_layout(guides = "collect", axes = "collect") |
| 172 | +``` |
| 173 | + |
| 174 | +## Controlling layout |
| 175 | + |
| 176 | +For more options for controlling layout, see the related vignette on the package website. |
| 177 | + |
| 178 | +- Adding empty areas |
| 179 | +- Inset plots |
| 180 | +- Adjusting widths and heights |
| 181 | + |
| 182 | +## Tags |
| 183 | + |
| 184 | +You can add "tags" to each panel with `plot_annotation()` |
| 185 | + |
| 186 | +```{r} |
| 187 | +p1 + p2 + plot_annotation(tag_levels = "A", tag_suffix = ")") |
| 188 | +``` |
| 189 | + |
| 190 | +## Modifying all panels |
| 191 | + |
| 192 | +You can use the `&` operator instead of `+` to modify **all** elements of a multi-panel figure. |
| 193 | + |
| 194 | +```{r} |
| 195 | +p1 + p2 & theme_bw() & scale_color_viridis_d() |
| 196 | +``` |
| 197 | + |
| 198 | +## Using with base R plots |
| 199 | + |
| 200 | +You can even combine `ggplot2` plots with base R plots with a special syntax |
| 201 | + |
| 202 | +```{r} |
| 203 | +p1 + ~hist(penguins$bill_depth_mm, main = "") |
| 204 | +``` |
| 205 | + |
| 206 | +## Getting Help |
| 207 | + |
| 208 | +- `patchwork`: [Package website](https://patchwork.data-imaginist.com/index.html) |
| 209 | + |
| 210 | +- `gganimate`: [Getting started guide](https://gganimate.com/articles/gganimate.html), [reference](https://gganimate.com/reference/index.html) |
| 211 | + |
| 212 | +- Our [drop-in hours](https://datascience.cct.arizona.edu/drop-in-hours) |
| 213 | + |
| 214 | +- [UA Data Science Slack](https://join.slack.com/t/uadatascience/shared_invite/zt-1xjxht9k0-E489WA6axO_SRVeKQh3cXg) |
| 215 | + |
| 216 | +# Anything we missed? |
| 217 | + |
| 218 | +Got a `ggplot2` question we didn't cover in this workshop series? |
| 219 | +Let's figure it out together! |
0 commit comments