Closed
Description
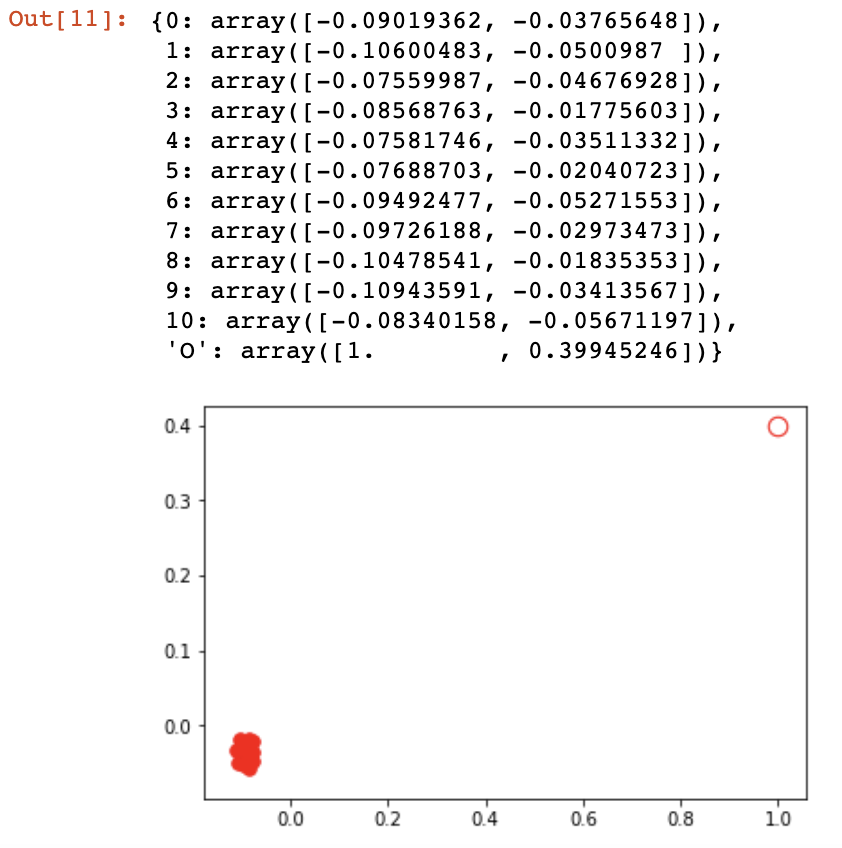
I'm trying to generate a diffnet in spring_layout using a 11x11 symmetrical sij with np.inf on diagonal:
sij_sym.npy.zip
%matplotlib inline
import matplotlib
from matplotlib import pyplot as plt
matplotlib.rcParams['font.size'] = 20
import networkx as nx
import numpy as np
from cvxopt import matrix
import diffnet as dn
import graph as gph
from netbfe import *
sij = matrix(np.sqrt(np.load("sij_sym.npy")))
#insert delta with stdev=1 for first s_ii:
delta = sij.shape[0] * [np.inf]
fict_delta = [1] + delta[1:]
#minimal graph generation:
n = networkBFEalloc( sij, N=1000, delta=fict_delta)
G = gph.diffnet_to_graph( n)
gph.draw_diffnet_graph( G, widthscale=1e-1, nodescale=5)
however this returns a graph where the origin node is distanced, skewing the network:
Metadata
Metadata
Assignees
Labels
No labels