diff --git a/01-intro.Rmd b/01-intro.Rmd
index a3ce4ab..d6f7a51 100644
--- a/01-intro.Rmd
+++ b/01-intro.Rmd
@@ -17,7 +17,7 @@ _This course is written for individuals who:_
- Have some familiarity with R or Python - have written some scripts.
- Have not had formal training in computational methods.
-- Have limited or no familiar with GitHub, Docker, or package management tools.
+- Have limited or no familiarity with GitHub, Docker, or package management tools.
```{r for_individuals_who, fig.alt="Intro to Reproducibility: For individuals who: Have some familiarity with R or Python - have written some scripts. Have not had formal training in computational methods. Have limited or no familiarity with GitHub. Advanced Reproducibility: For individuals who: Have completed the intro course and/or Have used GitHub somewhat.", out.width = "100%", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.g106226cdd08_0_0")
@@ -39,9 +39,9 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
Cancer datasets are plentiful, complicated, and hold untold amounts of information regarding cancer biology. Cancer researchers are working to apply their expertise to the analysis of these vast amounts of data but training opportunities to properly equip them in these efforts can be sparse. This includes training in reproducible data analysis methods.
-Data analyses are generally not reproducible without direct contact with the original researchers and a substantial amount of time and effort [@BeaulieuJones2017]. Reproducibility in cancer informatics (as with other fields) is still not monitored or incentivized despite that it is fundamental to the scientific method. Despite the lack of incentive, many researchers strive for reproducibility in their own work but often lack the skills or training to do so effectively.
+Data analyses are generally not reproducible without direct contact with the original researchers and a substantial amount of time and effort [@BeaulieuJones2017]. Reproducibility in cancer informatics (as with other fields) is still not monitored or incentivized despite the fact that it is fundamental to the scientific method. Even without incentives, most researchers strive for reproducibility in their own work but often lack the skills or training to do so effectively.
-Equipping researchers with the skills to create reproducible data analyses increases the efficiency of everyone involved. Reproducible analyses are more likely to be understood, applied, and replicated by others. This helps expedite the scientific process by helping researchers avoid false positive dead ends. Open source clarity in reproducible methods also saves researchers' time so they don't have to reinvent the proverbial wheel for methods that everyone in the field is already performing.
+Equipping researchers with the skills to create reproducible data analyses increases the efficiency of everyone involved. Reproducible analyses are more likely to be understood, applied, and replicated by others. This helps expedite the scientific process by helping researchers avoid false positive dead ends. Well documented and reproducible methods save researchers' time so they don't have to reinvent the proverbial wheel for methods that others in the field are already performing.
## Curriculum
@@ -51,13 +51,13 @@ This course introduces the concepts of reproducibility and replicability in the
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gd422c5de97_0_10")
```
-The course includes hands-on exercises for how to apply reproducible code concepts to their code. Individuals who take this course are encouraged to complete these activities as they follow along with the course material to help increase the reproducibility of their analyses.
+The course includes hands-on exercises for how to apply reproducible code concepts to your code. Individuals who take this course are encouraged to complete these activities as they follow along with the course material to help increase the reproducibility of their analyses.
**Goal of this course:**
-Equip learners with reproducibility skills they can apply to their existing analyses scripts and projects. This course opts for an "ease into it" approach. We attempt to give learners doable, incremental steps to increase the reproducibility of their analyses.
+Equip learners with reproducibility skills they can apply to their existing analysis scripts and projects. This course opts for an "ease into it" approach. We attempt to give learners doable, incremental steps to increase the reproducibility of their analyses.
**What is not the goal**
-This course is meant to introduce learners to the reproducibility tools, but _it does not necessarily represent the absolute end-all, be-all best practices for the use of these tools_. In other words, this course gives a starting point with these tools, but not an ending point. The advanced version of this course is the next step toward incrementally "better practices".
+This course is meant to introduce learners to reproducibility tools, but _it does not necessarily represent the absolute end-all, be-all best practices for the use of these tools_. In other words, this course gives a starting point with these tools, but not an ending point. The advanced version of this course is the next step toward incrementally "better practices".
```{r, fig.alt="Reproducibility is on a continuum. Goal of the course is to move learner’s skills toward creating reproducible analyses. This graph shows a two sided arrow with a gradient. On the very left is a ‘not repeatable analysis’ it was ran once. To the right of that is an analysis that ‘re-runs sometimes’. To the right of this, is an analysis that ‘Re-runs reliably in most contexts’. And all the way to the right is a ‘perfectly reproducible analysis’ that ‘Re-runs in every situation and gets the same result every time’. In red lettering we note that every analysis is started by being run once but no analysis is ‘perfectly reproducible’.", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.g1006ff8e7e9_48_3")
@@ -67,4 +67,4 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
This course is designed with busy professional learners in mind -- who may have to pick up and put down the course when their schedule allows.
-Each exercise has the option for you to continue along with the example files as you've been editing them in each chapter, OR you can download fresh chapter files that have been edited in accordance with the relative part of the course. This way, if you decide to skip a chapter or find that your own files you've been working on no longer make sense, you have a fresh starting point at each exercise.
+Each exercise has the option for you to continue along with the example files as you've been editing them in each chapter, OR you can download fresh chapter files that have been edited for that part of the course. This way, if you decide to skip a chapter or find that your own files no longer make sense, you have a fresh starting point for each exercise.
diff --git a/02-defining-reproducibility.Rmd b/02-defining-reproducibility.Rmd
index b4a28d9..fad786f 100644
--- a/02-defining-reproducibility.Rmd
+++ b/02-defining-reproducibility.Rmd
@@ -19,7 +19,7 @@ There's been a lot of discussion about what is included in the term `reproducibi
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_146")
```
-Reproducibility is related to repeatability and replicability but it is worth taking time to differentiate these terms
+Reproducibility is related to repeatability and replicability but it is worth taking time to differentiate these terms.
Perhaps you are like Ruby and have just found an interesting pattern through your data analysis! This has probably been the result of many months or years on your project and it's worth celebrating!
@@ -39,25 +39,25 @@ Given that Ruby's analysis is repeatable; she may feel confident now to share he
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_368")
```
-If Ruby's results are able to be reproduced by Avi, now Avi may collect new data and use Ruby's same analysis methods to analyze his data. Whether or not Avi's new data and results concur with Ruby's study's original inferences is known as _replicability_.
+If Ruby's results can be reproduced by Avi, now Avi may collect new data and use Ruby's same analysis methods to analyze his data. Whether or not Avi's new data and results concur with Ruby's study's original inferences is known as _replicability_.
```{r, fig.align='center', echo = FALSE, fig.alt= "Replicable: with new data do we obtain the same inferences? Ruby the researcher sends her code to Avi the Associate who then tries to re-run the same analysis on his own computer but now with new data (represented by a different color flashdrive). Will Avi come to the same inference that Ruby’s analysis did?"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_464")
```
-You may realize that these levels of research build on each other (like science is supposed to do). In this way, we can think of these in a hierarchy. Skipping any of these levels of research applicability can lead to unreliable results and conclusions.
+You may realize that these levels of research build on each other (like science is supposed to do). In this way, we can think of them in a hierarchy. Skipping any of these levels of research applicability can lead to unreliable results and conclusions.
```{r, fig.align='center', echo = FALSE, fig.alt= "A triangular graph shows a hierarchy of research. Repeatability is a the bottom ‘same researcher, same machine’, Runnability is next ‘same researcher, new machine’, Reproducibility is above that, ‘new researcher, same data’ and on the very top is Replicability ‘new researcher, new data’."}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1cd772e00_0_10")
```
-Science progresses when data and hypotheses are put through these levels thoroughly and sequentially. If results are not repeatable, they won't be reproducible or replicable.
+Science progresses when data and hypotheses are put through these levels thoroughly and sequentially. If results are not repeatable, they won't be reproducible or replicable. If they are not reproducible, they won't be replicable.
```{r, fig.align='center', echo = FALSE, fig.alt= "If your results are not repeatable they will NOT be reproducible. In other words, if you can’t get the same answer twice, other researchers won’t be able to get your answer either."}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_564")
```
-Ideally all analyses and results would be reproducible without too much time and effort spent; this would aid in the efficiency of research getting to the next stages and questions. But unfortunately, in practice, reproducibility is not as commonplace as we would hope. Institutions and reward systems generally do not prioritize or even measure reproducibility standards in research and training opportunities for reproducible techniques can be scarce. Reproducible research can often feel like an uphill battle that is made steeper by lack of training opportunities.
+Ideally all analyses and results would be reproducible without too much time and effort spent; this would aid in the efficiency of research getting to the next stages and questions. But unfortunately, in practice, reproducibility is not as commonplace as we would hope. Institutions and reward systems generally do not prioritize or even measure reproducibility in research, and training opportunities for reproducible techniques can be scarce. Reproducible research can often feel like an uphill battle that is made steeper by lack of training opportunities.
In this course, we hope to equip your research with the tools you need to enhance the reproducibility of your analyses so this uphill battle is _less steep_.
@@ -71,7 +71,7 @@ Let's say Ruby's results are repeatable in her own hands and she excitedly tells
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_179")
```
-Avi is also interested in Ruby's analysis methods and results. So Ruby sends Avi the code and data she used to obtain the results. Now, whether or not Avi is able to obtain the same exact results with this same data and same analysis code will indicate if Ruby's analysis is reproducible.
+Ruby sends Avi the code and data she used to obtain the results. Now, whether or not Avi is able to obtain the same exact results with this same data and same analysis code will indicate if Ruby's analysis is reproducible.
```{r, fig.align='center', echo = FALSE, fig.alt= "Ruby the researcher has her computer with a plot and a significant and exciting research result. Ruby says 'Here, Avi, this code runs well on my computer, let me email it to you!' Avi the associate says 'so exciting'"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_944")
@@ -79,13 +79,13 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
Ruby may have spent a lot of time on her code and getting it to work on her computer, but whether it will successfully run on Avi's computer is another story. Often when researchers share their analysis code it leads to a substantial amount of effort on the part of the researcher who has received the code to get it working and this often cannot be done successfully without help from the original code author [@BeaulieuJones2017].
-Avi is encountering errors because Ruby's code was written with Ruby's computer and local setup in mind and she didn't know how to make it more generally applicable. Avi is spending a lot of time just trying to re-run Ruby's same analysis on her same data; he has yet to be able to try the code on any additional data (which will likely bring up even more errors).
+Avi is encountering errors because Ruby's code was written with Ruby's computer and local setup in mind and she didn't know how to make it more generally usable. Avi is spending a lot of time just trying to re-run Ruby's same analysis on her same data; he has yet to be able to try the code on any additional data (which may bring up even more errors).
```{r, fig.align='center', echo = FALSE, fig.alt= "Avi the associate is confused and sweating. His computer has the word ‘error’ written all over it and its on fire trying to use Ruby’s code on Ruby’s data. This is using a substantial amount of time and effort on Avi’s part. "}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_993")
```
-Avi is still struggling to work with Ruby's code and is confused about the goals and approaches the code is taking. After struggling with Avi's code for an untold amount of time, Avi may decide it's time to email Ruby to get some clarity. Now both Avi and Ruby are confused about why this analysis isn't nicely re-running for Avi. Their attempts to communicate about the code through email haven't helped them clarify anything. Multiple versions of the code may have been sent back and forth between them and now things are taking a lot more time than either of them expected.
+Avi is still struggling to work with Ruby's code and is confused about the goals and approaches the code is taking. After struggling with Ruby's code for an untold amount of time, Avi may decide it's time to email Ruby to get some clarity. Now both Avi and Ruby are confused about why this analysis isn't nicely re-running for Avi. Their attempts to communicate about the code through email haven't helped them clarify anything. Multiple versions of the code may have been sent back and forth between them and now things are taking a lot more time than either of them expected.
```{r, fig.align='center', echo = FALSE, fig.alt= "Ruby the researcher and Avi are emailing back and forth. Avi is very confused about Ruby’s code and data. Avi’s computer says 'Error: file path 'Ruby’s computer/Ruby’s file/final_version10.R' not found'. Avi email says ‘Re:Re:Re: Data Hi Ruby, I don’t understand what this code is supposed to be doing…’ Ruby is also confused and emails back to Avi 'Re:Re:Re: Data Hi Avi, It works for me?''"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_1066")
@@ -103,7 +103,7 @@ Lack of errors also doesn't mean that either Ruby or Avi's runs of the code ran
Perhaps you've found yourself in a situation like Ruby and Avi; struggling to re-run code that you thought for sure was working a minute ago. In the upcoming chapters, we will discuss how to bolster your projects' reproducibility.
-As you apply these reproducible techniques to your own projects, you may feel like it is taking more time to reach endpoints, but keep in mind that reproducible analyses and projects have higher upfront costs but these will absolutely pay off in the long term.
+As you apply these reproducible techniques to your own projects, you may feel like it is taking more time to reach endpoints, but keep in mind that reproducible analyses and projects have higher upfront costs but will absolutely pay off in the long term.
```{r, fig.align='center', echo = FALSE, fig.alt= "Reproducibility is a tortoise’s game - it’s an incremental and slow process but it has high payoffs!"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf7bed24491_1_38")
@@ -115,7 +115,7 @@ Reproducibility in your analyses is not only a time saver for yourself, but also
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1cd772e00_0_5")
```
-You might not change a single character in your code but then return to it in a a few days/months/years and find that it no longer runs! Reproducible code stands the test of time longer, making 'future you' glad you spent the time to work on it. It's said that your closest collaborator is you from 6 months ago but you don't reply to email [@Broman].
+You might not change a single character in your code but then return to it in a few days/months/years and find that it no longer runs! Reproducible code stands the test of time longer, making 'future you' glad you spent the time to work on it. It's said that your closest collaborator is you from 6 months ago but you don't reply to email [@Broman].
```{r, fig.align='center', echo = FALSE, fig.alt= "Ruby the researcher’s code works now as represented on her computer by a check mark. But Future Ruby, who has gray hair has an error running the same code!"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1cd772e00_0_330")
@@ -125,7 +125,7 @@ Many a data scientist has referred to their frustration with their past selves:
-The more you comment your code, and make it clear and readable, your future self will thank you.
+The more you comment your code, and make it clear and readable, the more your future self will thank you.
Reproducible code also saves your colleagues time! The more reproducible your code is, the less time all of your collaborators will need to spend troubleshooting it. The more people who use your code and need to try to fix it, the more time is wasted. This can add up to a lot of wasted researcher time and effort.
@@ -142,7 +142,7 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
## Reproducibility exists on a continuum!
-Incremental work on your analyses is good! You do not need to make your analyses perfect on the first try or even within a particular time frame. The first step in creating an analysis is to get it to work once! But the work does not end there. Furthermore, no analysis is or will ever be perfect in that it will not be reproducible in every single context throughout time. incrementally pushing our analyses toward the right of this continuum is the goal.
+Incremental work on your analyses is good! You do not need to make your analyses perfect on the first try or even within a particular time frame. The first step in creating an analysis is to get it to work once! But the work does not end there. Furthermore, no analysis is or will ever be perfect in that it will not be reproducible in every single context throughout time. Incrementally pushing our analyses toward the right of this continuum is the goal.
```{r, fig.align='center', echo = FALSE, fig.alt= "Reproducibility is on a continuum. This graph shows a two sided arrow with a gradient. On the very left is a ‘not repeatable analysis’ it was ran once. To the right of that is an analysis that ‘re-runs sometimes’. To the right of this, is an analysis that ‘Re-runs reliably in most contexts’. And all the way to the right is a ‘perfectly reproducible analysis’ that ‘Re-runs in every situation and gets the same result every time’. In red lettering we note that every analysis is started by being run once but no analysis is ‘perfectly reproducible’."}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf7bed24491_1_0")
diff --git a/03-project-organization.Rmd b/03-project-organization.Rmd
index 73d4099..92abf2c 100644
--- a/03-project-organization.Rmd
+++ b/03-project-organization.Rmd
@@ -17,7 +17,7 @@ Keeping your files organized is a skill that has a high long-term payoff. As you
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf7bed24491_1_56")
```
-@Tayo2019 discusses four particular reasons why it is important to organize your project:
+@Tayo2019 discusses four reasons why it is important to organize your project:
> 1. Organization increases productivity. If a project is well organized, with everything placed in one directory, it makes it easier to avoid wasting time searching for project files such as datasets, codes, output files, and so on.
> 2. A well-organized project helps you to keep and maintain a record of your ongoing and completed data science projects.
@@ -48,8 +48,8 @@ Getting more specific, here's some ideas of how to organize your project:
- **Make file names informative** to those who don't have knowledge of the project but avoid using spaces, quotes, or unusual characters in your filenames and folders -- these only serve to make reading in files a nightmare in some programs.
- **Number scripts** in the order that they are run.
-- **Keep like-files together** in their own directory: results tables with other results tables, etc. _Including most importantly keeping raw data separate from processed data or other results!_
-- **Put source scripts and functions in their own directory**. Things that should never need to be called directly by yourself or anyone else.
+- **Keep like-files together** in their own directory: results tables with other results tables, etc. _Most importantly, keep raw data separate from processed data or other results!_
+- **Put source scripts and functions in their own directory**. These are things that are called directly by yourself or anyone else.
- **Put output in its own directories** like `results` and `plots`.
- **Have a central document (like a README)** that describes the basic information about the analysis and how to re-run it.
- Make it easy on yourself, **dates aren't necessary**. The computer keeps track of those.
@@ -62,14 +62,14 @@ Let's see what these principles might look like put into practice.
Here's an example of what this might look like:
```
project-name/
-├── run_analysis.sh
+├── run-analysis.sh
├── 00-download-data.sh
├── 01-make-heatmap.Rmd
├── README.md
├── plots/
│ └── project-name-heatmap.png
├── results/
-│ └── top_gene_results.tsv
+│ └── top-gene-results.tsv
├── raw-data/
│ ├── project-name-raw.tsv
│ └── project-name-metadata.tsv
@@ -82,14 +82,14 @@ project-name/
**What these hypothetical files and folders contain:**
-- `run_analysis.sh` - A central script that runs everything again
-- `00-download-data.sh` - The script that needs to be run first and is called by run_analysis.sh
-- `01-make-heatmap.Rmd` - The script that needs to be run second and is also called by run_analysis.sh
-- `README.md` - The document that has the information that will orient someone to this project, we'll discuss more about how to create a helpful README in [an upcoming chapter](https://jhudatascience.org/Reproducibility_in_Cancer_Informatics/documenting-analyses.html#readmes).
+- `run-analysis.sh` - A central script that runs everything again
+- `00-download-data.sh` - The script that needs to be run first and is called by run-analysis.sh
+- `01-make-heatmap.Rmd` - The script that needs to be run second and is also called by run-analysis.sh
+- `README.md` - A document that will orient someone to this project. We'll discuss more about how to create a helpful README in [an upcoming chapter](https://jhudatascience.org/Reproducibility_in_Cancer_Informatics/documenting-analyses.html#readmes).
- `plots` - A folder of plots and resulting images
- `results` - A folder results
-- `raw-data` - Data files as they first arrive and **nothing** has been done to them yet.
-- `processed-data` - Data that has been modified from the raw in some way.
+- `raw-data` - Data files as they first arrive (**nothing** has been done to them yet)
+- `processed-data` - Data that has been modified from their raw form in some way
- `util` - A folder of utilities that never needs to be called or touched directly unless troubleshooting something
## Readings about organizational strategies for data science projects:
@@ -99,13 +99,13 @@ You can read through some of these articles to think about what kind of organiza
- [Jenny Bryan's organizational strategies](https://www.stat.ubc.ca/~jenny/STAT545A/block19_codeFormattingOrganization.html) [@Bryan2021].
- [Danielle Navarro's organizational strategies](https://www.youtube.com/playlist?list=PLRPB0ZzEYegPiBteC2dRn95TX9YefYFyy) @Navarro2021
-- [Jenny Bryan on Project-oriented workflows](https://www.tidyverse.org/blog/2017/12/workflow-vs-script/)[@Bryan2017].
+- [Jenny Bryan on Project-oriented workflows](https://www.tidyverse.org/blog/2017/12/workflow-vs-script/) [@Bryan2017].
- [Data Carpentry mini-course about organizing projects](https://datacarpentry.org/organization-genomics/) [@DataCarpentry2021].
- [Andrew Severin's strategy for organization](https://bioinformaticsworkbook.org/projectManagement/Intro_projectManagement.html#gsc.tab=0) [@Severin2021].
- [A BioStars thread where many individuals share their own organizational strategies](https://www.biostars.org/p/821/) [@Biostars2021].
- [Data Carpentry course chapter about getting organized](https://bioinformatics-core-shared-training.github.io/shell-genomics/07-organization/index.html) [@DataCarpentry2019].
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## Get the exercise project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-3.zip).
@@ -141,8 +141,8 @@ unzip -o chapter-zips/r-heatmap-chapt-3.zip -d chapter-zips/
Using your computer's GUI (drag, drop, and clicking), organize the files that are part of this project.
-1. Organized these files using an organizational scheme similar to [what is described above](#example organizational-scheme).
-1. Create folders like `plots`, `results`, and `data` folder. Note that `aggregated_metadata.json` and `LICENSE.TXT` also belong in the `data` folder.
+1. Organize these files using an organizational scheme similar to [what is described above](#example-organizational-scheme).
+1. Create folders like `plots`, `results`, and `data`. Note that `aggregated_metadata.json` and `LICENSE.TXT` also belong in the `data` folder.
1. You will want to delete any files that say "OLD". Keeping multiple versions of your scripts around is a recipe for mistakes and confusion. In the advanced course we will discuss how to use version control to help you track this more elegantly.
After your files are organized, you are ready to move on to the next chapter and create a notebook!
diff --git a/04-open-source-with-github.Rmd b/04-open-source-with-github.Rmd
index 749f86b..5efafc0 100644
--- a/04-open-source-with-github.Rmd
+++ b/04-open-source-with-github.Rmd
@@ -42,7 +42,7 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
#### Keep a documented history of your project
-Overtime in a project, a lot happens, especially when it comes to exploring and handling data. Sometimes the rationale behind decisions that were made around an analysis can get lost. GitHub keeps communications and tracks the changes to your files so that you don't have to revisit a question you already answered.
+Over time in a project, a lot happens, especially when it comes to exploring and handling data. Sometimes the rationale behind decisions that were made around an analysis can get lost. GitHub keeps communications and tracks the changes to your files so that you don't have to revisit a question you already answered.
```{r, fig.alt="Ruby holds a magnifying glass and says 'Why did we write the code this way? I don’t remember… Good thing through git tracking I can look into this file’s history and remind myself how it became this.'", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf62875ddf7_0_92")
@@ -58,13 +58,13 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
#### Experiment with your analysis
-Data science projects often lead to side analyses that could be very worth while but might be scary to venture on if you don't have your code well version controlled. Git and GitHub allow you to venture on these side experiments without fear since your main code can be kept safe from your side venture.
+Data science projects often lead to side analyses that could be very worthwhile but might be scary to venture on if you don't have your code well version controlled. Git and GitHub allow you to venture on these side experiments without fear since your main code can be kept safe from your side venture.
```{r, fig.alt="Ruby says ‘I’m not sure if this side analysis I’m working on is a good idea or not, but I want to test it. Good thing I can make a separate branch and keep my original code safe from my experimenting.’ Her computer shows her main code and a branch off of it that says ‘test analysis’. After time and work goes by she may decide to incorporate her test analysis with her main code", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf62875ddf7_0_47")
```
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## Get the exercise project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-4.zip).
@@ -101,7 +101,7 @@ unzip -o chapter-zips/r-heatmap-chapt-4.zip -d chapter-zips/
Now that we understand how useful GitHub is for creating reproducible analyses, it's time to set ourselves up on GitHub.
-Git and GitHub have a whole rich world of tools and terms that can get complex quickly, but for this exercise, we will not worry about those terms and functionalities just yet, but focus on getting code up on GitHub so we are ready to collaborate and conduct open analyses!
+Git and GitHub have a whole rich world of tools and terms that can get complex quickly, but for this exercise, we will not worry about those terms and functionalities just yet, and instead focus on getting code up on GitHub so we are ready to collaborate and conduct open analyses!
- Go to Github's main page and click Sign Up if you don't have an account.
- Follow these instructions to create a repository. As a general, but not absolute rule, you will want to keep one GitHub repository for one analysis project.
diff --git a/05-scientific-notebooks.Rmd b/05-scientific-notebooks.Rmd
index 34162ce..7ebb9da 100644
--- a/05-scientific-notebooks.Rmd
+++ b/05-scientific-notebooks.Rmd
@@ -50,20 +50,20 @@ This can also be handy especially if you use automation to enhance the reproduci
Because of all of these reasons, we encourage the use of computational notebooks as a means of enhancing reproducibility. (This course itself is also written with the use of notebooks!)
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## Get the exercise project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-5.zip).
```{bash, include = FALSE}
-mkdir -p chapter-zips
-wget -O chapter-zips/python-heatmap-chapt-5.zip https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-5.zip
+#mkdir -p chapter-zips
+#wget -O chapter-zips/python-heatmap-chapt-5.zip https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-5.zip
```
Now double click your chapter zip file to unzip. For Windows you may have to [follow these instructions](https://support.microsoft.com/en-us/windows/zip-and-unzip-files-f6dde0a7-0fec-8294-e1d3-703ed85e7ebc).
```{bash, include = FALSE}
-unzip -o chapter-zips/python-heatmap-chapt-5.zip -d chapter-zips/
+#unzip -o chapter-zips/python-heatmap-chapt-5.zip -d chapter-zips/
```
@@ -71,14 +71,14 @@ unzip -o chapter-zips/python-heatmap-chapt-5.zip -d chapter-zips/
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/r-heatmap-chapt-5.zip).
```{bash, include = FALSE}
-mkdir -p chapter-zips
-wget -O chapter-zips/r-heatmap-chapt-5.zip https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/r-heatmap-chapt-5.zip
+#mkdir -p chapter-zips
+#wget -O chapter-zips/r-heatmap-chapt-5.zip https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/r-heatmap-chapt-5.zip
```
Now double click your chapter zip file to unzip. For Windows you may have to [follow these instructions](https://support.microsoft.com/en-us/windows/zip-and-unzip-files-f6dde0a7-0fec-8294-e1d3-703ed85e7ebc).
```{bash, include = FALSE}
-unzip -o chapter-zips/r-heatmap-chapt-5.zip -d chapter-zips/
+#unzip -o chapter-zips/r-heatmap-chapt-5.zip -d chapter-zips/
```
@@ -127,7 +127,7 @@ For this chapter, we will create notebooks from our example files code. Notebook
**Getting familiar with RStudio's interface**
- +
+ > The RStudio environment has four main **panes**, each of which may have a number of tabs that display different information or functionality. (their specific location can be changed under Tools -> Global Options -> Pane Layout).
@@ -165,11 +165,13 @@ Now, in your respective IDE, we'll turn our unreproducible scripts into notebook
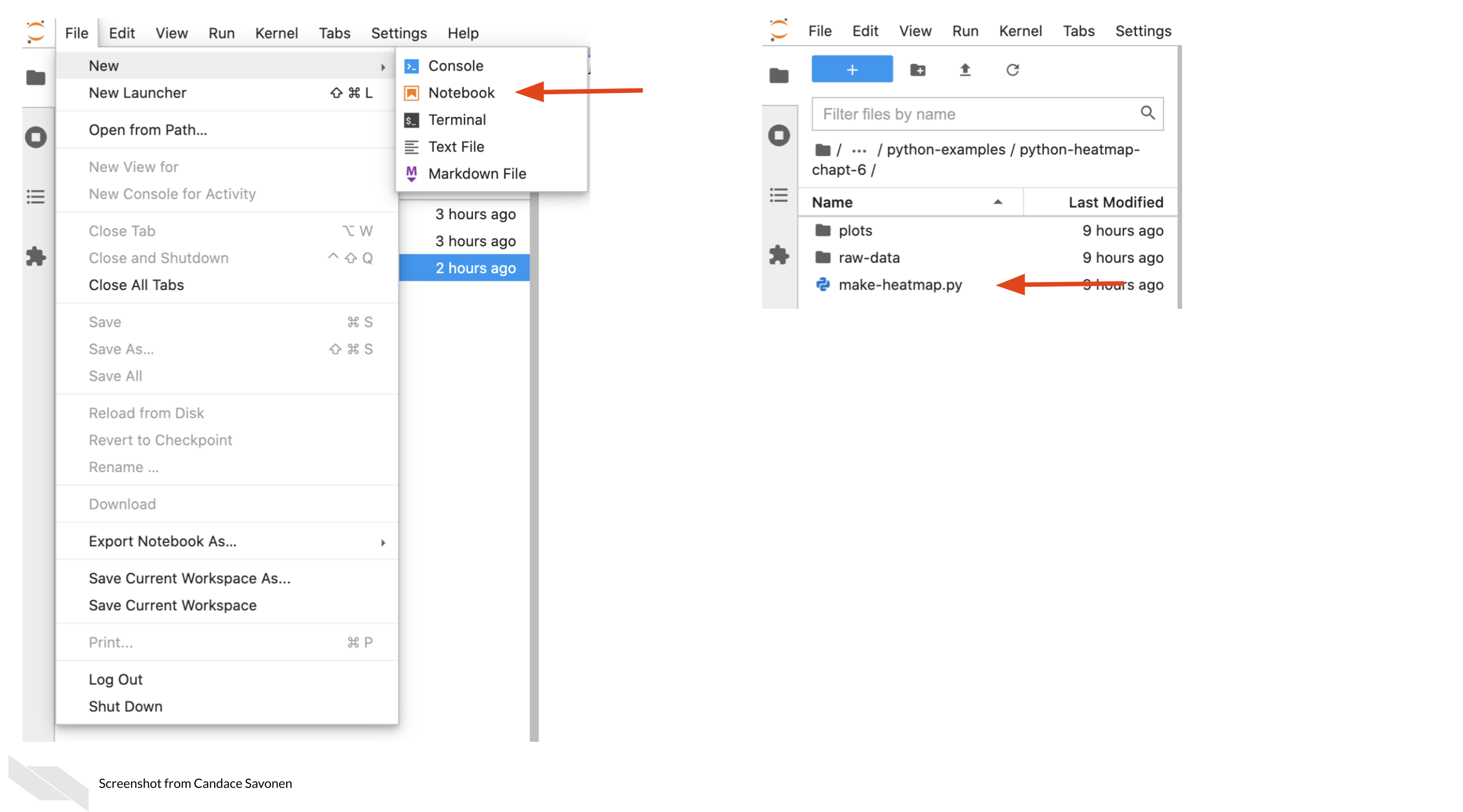
1. Start a new notebook by going to `New` > `Notebook`.
2. Then open up this chapter’s example code folder and open the `make-heatmap.py` file.
-
> The RStudio environment has four main **panes**, each of which may have a number of tabs that display different information or functionality. (their specific location can be changed under Tools -> Global Options -> Pane Layout).
@@ -165,11 +165,13 @@ Now, in your respective IDE, we'll turn our unreproducible scripts into notebook
1. Start a new notebook by going to `New` > `Notebook`.
2. Then open up this chapter’s example code folder and open the `make-heatmap.py` file.
- +
+ +
3. Create a new code chunk in your notebook.
-
+
3. Create a new code chunk in your notebook.
- +
+
+
+ 4. Now copy and paste all of the code from `make-heatmap.py` into a new chunk. We will later break up this large chunk of code into smaller chunks that are thematic in the next chapter.
@@ -181,16 +183,20 @@ For [more about using Jupyter notebooks see this](https://realpython.com/jupyter
4. Now copy and paste all of the code from `make-heatmap.py` into a new chunk. We will later break up this large chunk of code into smaller chunks that are thematic in the next chapter.
@@ -181,16 +183,20 @@ For [more about using Jupyter notebooks see this](https://realpython.com/jupyter
**Set up an R notebook**
-1. Start a new notebook by going to `File` > `New Files` > `R Notebook`.
+1. Start a new notebook by going to `File` > `New File` > `R Notebook`.
+
+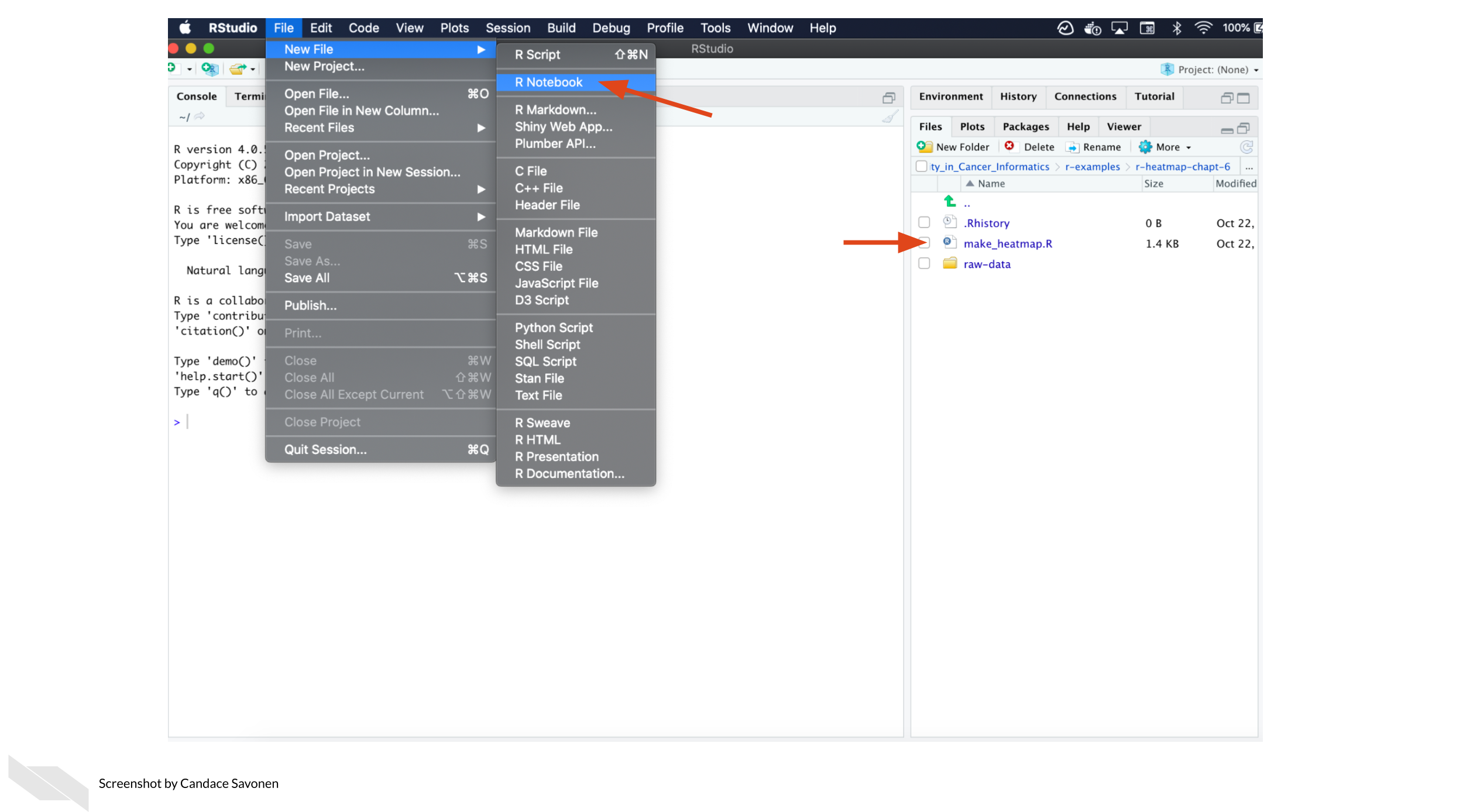 +
2. Then open up this chapter’s example code folder and open the `make_heatmap.R` file.
-
+
2. Then open up this chapter’s example code folder and open the `make_heatmap.R` file.
- 3. Practice creating a new chunk in your R notebook by clicking the `Code` > `Insert Chunk` button on the toolbar or by pressing `Cmd+Option+I` (in Mac) or `Ctrl + Alt + I` (in Windows). (You can also manually type out the back ticks and `{}`)
4. Delete all the default text in this notebook but keep the header which is surrounded by `---` and looks like:
+
`````
title: "R Notebook"
output: html_notebook
`````
-You can feel free to change the title from `R Notebook` to something that better suits the contents of this notebook.
+You can feel free to change the title from `R Notebook` to something that better suits the contents of this notebook.
+
5. Now copy and paste all of the code from `make_heatmap.R` into a new chunk. We will later break up this large chunk of code into smaller chunks that are thematic in the next chapter.
6. Save your `untitled.Rmd` into something that tells us what it will end up doing like `make-heatmap.Rmd`.
7. Notice that upon saving your `.Rmd` file, a new file `.nb.html` file of the same name is created. Open that file and choose `view in Browser`. If RStudio asks you to choose a browser, then choose a default browser.
@@ -198,8 +204,15 @@ You can feel free to change the title from `R Notebook` to something that better
For [more about using R notebooks see this](https://bookdown.org/yihui/rmarkdown/) by @Xie2018.
+
+
+
3. Practice creating a new chunk in your R notebook by clicking the `Code` > `Insert Chunk` button on the toolbar or by pressing `Cmd+Option+I` (in Mac) or `Ctrl + Alt + I` (in Windows). (You can also manually type out the back ticks and `{}`)
4. Delete all the default text in this notebook but keep the header which is surrounded by `---` and looks like:
+
`````
title: "R Notebook"
output: html_notebook
`````
-You can feel free to change the title from `R Notebook` to something that better suits the contents of this notebook.
+You can feel free to change the title from `R Notebook` to something that better suits the contents of this notebook.
+
5. Now copy and paste all of the code from `make_heatmap.R` into a new chunk. We will later break up this large chunk of code into smaller chunks that are thematic in the next chapter.
6. Save your `untitled.Rmd` into something that tells us what it will end up doing like `make-heatmap.Rmd`.
7. Notice that upon saving your `.Rmd` file, a new file `.nb.html` file of the same name is created. Open that file and choose `view in Browser`. If RStudio asks you to choose a browser, then choose a default browser.
@@ -198,8 +204,15 @@ You can feel free to change the title from `R Notebook` to something that better
For [more about using R notebooks see this](https://bookdown.org/yihui/rmarkdown/) by @Xie2018.
+
+
+
+
+
+
+
Now that you've created your notebook, you are ready to start polishing that code!
**Any feedback you have regarding this exercise is greatly appreciated; you can fill out [this form](https://forms.gle/ygSSwoGaEATA2S65A)!**
diff --git a/06-package-management.Rmd b/06-package-management.Rmd
index f0e3563..bae1e42 100644
--- a/06-package-management.Rmd
+++ b/06-package-management.Rmd
@@ -17,7 +17,7 @@ As we discussed previously, sometimes two different researchers can run the same
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf1accd298e_0_673")
```
-What Ruby and Avi may not realize is that although they may have used the same code and data, the software packages that they have on each of their computers might be very different. Even if they have the same software packages, they likely don't have the same versions and versions can influence results! Different computing environments are not only a headache to detangle, they also can influence the reproducibility of your results [ @BeaulieuJones2017].
+What Ruby and Avi may not realize is that although they have used the same code and data, the software packages that they have on each of their computers may be very different. Even if they have the same software packages, they likely don't have the same package versions and these versions can influence results! Different computing environments are not only a headache to detangle, they also can influence the reproducibility of your results [ @BeaulieuJones2017].
```{r, fig.alt="Ruby has a particular computing environment she has developed her code from. This computing environment is represented as a bubble above her computer with various hexagons with version numbers as well as Rstudio and R installed on her computer. Her code ran just fine on her particular computing environment. Avi attempted to run Ruby’s code on his very different local computing environment and got an error. His computer runs the same code but came up with a different result!", out.width = "100%", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf62875ddf7_0_404")
@@ -29,11 +29,11 @@ There are two strategies for dealing with software versions that we will discuss
### Strategy 1: Session Info - record a list of your packages
-One strategy to combat different software versions is to list the **session info**. This is the easiest (though not most comprehensive) method for handling differences in software versions is to have your code list details about your computing environment.
+One strategy to combat different software versions is to list the **session info**. The easiest (though not most comprehensive) method for handling differences in software versions is to have your code list details about your computing environment.
-Session info can lead to clues as to why results weren't reproducible. For example, if both Avi and Ruby ran notebooks and included a session info print out it may look like this:
+Session info can lead to clues as to why results weren't reproducible. For example, if both Avi and Ruby ran notebooks, and included a session info print out, it may look like this:
-```{r, fig.alt="Two session info print outs are show side by side. One is labeled as ‘Ruby’s session info print out’ and the other as ‘Avi’s session info print out’. Highlighted we can see that they have different R versions: 4.0.2 vs 4.0.5. They also have different operating systems. The packages they have attached is rmarkdown but they also have different rmarkdown package versions! If Avi and Ruby have discrepancies in their results, the session info print out gives a record which may have clues to why that might be! This can give them items to look into for determining why the results didn’t reproduce as expected.", out.width = "100%", echo = FALSE}
+```{r, fig.alt="Two session info print outs are show side by side. One is labeled as ‘Ruby’s session info print out’ and the other as ‘Avi’s session info print out’. Highlighted we can see that they have different R versions: 4.0.2 vs 4.0.5. They also have different operating systems. The packages they have attached is rmarkdown but they also have different rmarkdown package versions! If Avi and Ruby have discrepancies in their results, the session info print out gives a record which may have clues as to why that might be! This can suggest items to look into for determining why the results didn’t reproduce as expected.", out.width = "100%", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.g102dc56db08_0_0")
```
@@ -41,7 +41,7 @@ Session info shows us that they have different R versions and different operatin
### Strategy 2: Package managers - share a useable snapshot of your environment
-**Package managers** can help handle your computing environment for you in a way that you can share them with others. In general, package managers work by capturing a snapshot of the environment and when that environment snapshot is shared, it attempt to rebuild it. For R and Python versions of the exercises, we will be using different managers, but the foundational strategy will be the same: include a file that someone else could replicate your package set up from.
+**Package managers** can help handle your computing environment for you in a way so that you can share it with others. In general, package managers work by capturing a snapshot of the environment and when that environment snapshot is shared, it attempts to rebuild it. For R and Python versions of the exercises, we will be using different managers, but the foundational strategy will be the same: include a file that someone else could replicate your package setup from.
```{r, fig.alt="In general, package managers work by capturing a snapshot of the environment and when that environment snapshot is shared, it attempt to rebuild it. In this example we show that Ruby has an environment, and using a package manager, has taken a snapshot of her computing environment. That snapshot is shared with Avi, who can use the package manager to attempt to build it on his own computer. This will help address some differences in package versions between two individual’s computers.", out.width = "100%", echo = FALSE}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.g102dc56db08_49_26")
@@ -52,7 +52,7 @@ For both exercises, we will download an environment 'snapshot' file we've set up
- For Python, we'll use `conda` for package management and store this information in a `environment.yml` file.
- For R, we'll use `renv` for package management and store this information in a `renv.lock` file.
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## Get the exercise project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-6.zip).
@@ -98,11 +98,16 @@ In your scientific notebook, you'll need to add two items.
**R version of the exercise**
1. In your Rmd file, add a chunk in the very end that looks like this:
-`````
-```{r}
+
+```{r, eval = FALSE}
+sessionInfo()
+```
+
+The output will look something like this:
+
+```{r, echo = FALSE}
sessionInfo()
```
-`````
2. Save your notebook as is. Note it will not run correctly until we address the issues with the code in the next chapter.
@@ -151,7 +156,7 @@ jupyter lab
1. Go to RStudio and the Console pane:
-2. Install `renv` using (you should only need to do this once per your computer or RStudio environment).
+2. Install `renv` using the command below (you should only need to do this once per your computer or RStudio environment).
```
install.packages("renv")
diff --git a/07-durable-code.Rmd b/07-durable-code.Rmd
index 9e79248..c64a40e 100644
--- a/07-durable-code.Rmd
+++ b/07-durable-code.Rmd
@@ -17,12 +17,12 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
 Getting your code to work the first time is the first step, but don't stop there!
-Just like in writing a manuscript you wouldn't consider your first draft a final draft, your polishing code works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
+Just like in writing a manuscript you wouldn't consider your first draft a final draft, your code polishing works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
#### Prioritize readability over cleverness
Getting your code to work the first time is the first step, but don't stop there!
-Just like in writing a manuscript you wouldn't consider your first draft a final draft, your polishing code works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
+Just like in writing a manuscript you wouldn't consider your first draft a final draft, your code polishing works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
#### Prioritize readability over cleverness
 -Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness comprises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, including your future self!).
+Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness compromises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, this includes your future self!).
What does readable code look like? @Orosz2019 has some thoughts on writing readable code:
@@ -34,7 +34,7 @@ What does readable code look like? @Orosz2019 has some thoughts on writing reada
> **The real test of readable code is others reading it.** So get feedback from others, via code reviews. Ask people to share feedback on how clear the code is. Encourage people to ask questions if something does not make sense. Code reviews - especially thorough code reviews - are the best way to get feedback on how good and readable your code is.
>
-> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints to opportunities to make the code more readable.
+> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints at opportunities to make the code more readable.
>
> A good way to get more feedback on the clarity of your code is to ask for feedback from someone who is not an expert on the codebase you are working on. Ask specifically for feedback on how easy to read your code is. Because this developer is not an expert on the codebase, they'll focus on how much they can follow your code. Most of the comments they make will be about your code's readability.
@@ -60,7 +60,7 @@ If you find yourself writing something more than once, you might want to write a
DRY code is easier on the reviewer because they don't have to review the same thing twice, but also because they don't have to review the same thing twice. ;)
DRYing code is something that takes some iterative passes and edits through your code, but in the end DRY code saves you and your collaborators time and can be something you reuse again in a future project!
-Here's an slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
+Here's a slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
```
paste('Hello','John', 'welcome to this course')
@@ -109,9 +109,9 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
-Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness comprises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, including your future self!).
+Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness compromises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, this includes your future self!).
What does readable code look like? @Orosz2019 has some thoughts on writing readable code:
@@ -34,7 +34,7 @@ What does readable code look like? @Orosz2019 has some thoughts on writing reada
> **The real test of readable code is others reading it.** So get feedback from others, via code reviews. Ask people to share feedback on how clear the code is. Encourage people to ask questions if something does not make sense. Code reviews - especially thorough code reviews - are the best way to get feedback on how good and readable your code is.
>
-> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints to opportunities to make the code more readable.
+> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints at opportunities to make the code more readable.
>
> A good way to get more feedback on the clarity of your code is to ask for feedback from someone who is not an expert on the codebase you are working on. Ask specifically for feedback on how easy to read your code is. Because this developer is not an expert on the codebase, they'll focus on how much they can follow your code. Most of the comments they make will be about your code's readability.
@@ -60,7 +60,7 @@ If you find yourself writing something more than once, you might want to write a
DRY code is easier on the reviewer because they don't have to review the same thing twice, but also because they don't have to review the same thing twice. ;)
DRYing code is something that takes some iterative passes and edits through your code, but in the end DRY code saves you and your collaborators time and can be something you reuse again in a future project!
-Here's an slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
+Here's a slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
```
paste('Hello','John', 'welcome to this course')
@@ -109,9 +109,9 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
*Why do you need to refresh your kernel/session?*
-As a quick example of why refreshing your kernel/session, let's suppose you are troubleshooting something that centers around an object named `some_obj` but then you rename this object to `iris_df`. When you rename this object you may need to update this other places in the code. If you don't refresh your environment while working on your code, `some_obj` will still be in your environment. This will make it more difficult for you to find where else the code needs to be updated.
+As a quick example of why refreshing your kernel/session helps, let's suppose you are troubleshooting something that centers around an object named `some_obj` but then you rename this object to `iris_df`. When you rename this object you may need to update this other places in the code. If you don't refresh your environment while working on your code, `some_obj` will still be in your environment. This will make it more difficult for you to find where else the code needs to be updated.
-Refreshing your kernel/session goes beyond objects defined in your environment, and also can affect packages and dependencies loaded or all kinds of other things attached to your kernel/session.
+Refreshing your kernel/session goes beyond objects defined in your environment, and also can affect the packages and dependencies loaded, or all kinds of other things attached to your kernel/session.
As a quick experiment, try this in your Python or R environment:
@@ -185,7 +185,7 @@ Try to avoid using variable names that have no meaning like `tmp` or `x`, or `i`
> 2 Use consistent notation for naming convention.
> 3 Use standard terms.
> 4 Do not number a variable name.
-> 5 When you find another way to name variable, refactor as fast as possible.
+> 5 When you find another word or term to replace and improve your variable name, refactor those code as soon as possible.
[@Hobert2018]
@@ -198,9 +198,9 @@ Try to avoid using variable names that have no meaning like `tmp` or `x`, or `i`
#### Follow a code style
 -Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may even work all the same, just like you understood what I wrote in that last sentence, but a lack of consistent style can make require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
+Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may run correctly, just like you understood what I wrote in that last sentence, but a lack of consistent style can require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
-There are different style guides out there that people adhere to. It doesn't matter so much which one you choose, so much that you pick one and stick to it for a particular project.
+There are different style guides out there that people adhere to. It doesn't matter which one you choose, as long as you pick one and stick to it for a particular project.
_Python style guides_:
@@ -212,7 +212,7 @@ _R style guides_:
- [Hadley Wickham's Style guide](http://adv-r.had.co.nz/Style.html) @Wickham.
- [Google R style guide](https://google.github.io/styleguide/Rguide.html) @GoogleR.
-Although writing code following a style as you are writing is a good practice, we're all human and that can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
+Although writing code that follows a style is a good practice, we're all human and it can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
For Python code, you can use [python black](https://black.readthedocs.io/en/stable/) and for R, [styler](https://www.tidyverse.org/blog/2017/12/styler-1.0.0/).
#### Organize the structure of your code
@@ -304,7 +304,7 @@ There's so many opinions and strategies on best practices for code. And although
- [R for Epidemiology: Coding best practices](https://www.r4epi.com/chapters/best_practices/best_practices) by @Cannell2021.
- [Best practices for R Programming](https://towardsdatascience.com/best-practices-for-r-programming-ec0754010b5a) by @Bernardo2021.
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## See example project files
-Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may even work all the same, just like you understood what I wrote in that last sentence, but a lack of consistent style can make require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
+Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may run correctly, just like you understood what I wrote in that last sentence, but a lack of consistent style can require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
-There are different style guides out there that people adhere to. It doesn't matter so much which one you choose, so much that you pick one and stick to it for a particular project.
+There are different style guides out there that people adhere to. It doesn't matter which one you choose, as long as you pick one and stick to it for a particular project.
_Python style guides_:
@@ -212,7 +212,7 @@ _R style guides_:
- [Hadley Wickham's Style guide](http://adv-r.had.co.nz/Style.html) @Wickham.
- [Google R style guide](https://google.github.io/styleguide/Rguide.html) @GoogleR.
-Although writing code following a style as you are writing is a good practice, we're all human and that can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
+Although writing code that follows a style is a good practice, we're all human and it can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
For Python code, you can use [python black](https://black.readthedocs.io/en/stable/) and for R, [styler](https://www.tidyverse.org/blog/2017/12/styler-1.0.0/).
#### Organize the structure of your code
@@ -304,7 +304,7 @@ There's so many opinions and strategies on best practices for code. And although
- [R for Epidemiology: Coding best practices](https://www.r4epi.com/chapters/best_practices/best_practices) by @Cannell2021.
- [Best practices for R Programming](https://towardsdatascience.com/best-practices-for-r-programming-ec0754010b5a) by @Bernardo2021.
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## See example project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-7.zip).
@@ -358,9 +358,9 @@ For example, for this `make-heatmap` notebook we want to:
**The exercise: Polishing code**
-1. Start up JupyterLab with running `jupyter lab` from your command line.
+1. Start up JupyterLab by running `jupyter lab` from your command line.
2. Activate your conda environment using `conda activate reproducible-python`.
-3. Open up your notebook you made in the previous chapter `make-heatmap.ipynb`
+3. Open up the notebook you made in the previous chapter `make-heatmap.ipynb`
4. Work on organizing the code chunks and adding documentation to reflect the steps we've laid out in the [previous section](#organize-the-big-picture-of-the-code), you may want to work on this iteratively as we dive into the code.
5. As you clean up the code, you should run and re-run chunks to see if they work as you expect. You will also want to refresh your environment to help you develop the code (sometimes older objectives stuck in your environment can inhibit your ability to troubleshoot). In Jupyter, you refresh your environment by using the `refresh` icon in the toolbar or by going to `Restart Kernel`.
@@ -489,7 +489,7 @@ _More reading on the tidyverse:_
2. Open up the notebook you created in the previous chapter.
3. Now we'll work on applying the principles from this chapter to the code. We'll cover some of the points here, but then we encourage you to dig into the fully transformed notebook we will link at the end of this section.
4. Work on organizing the code chunks and adding documentation to reflect the steps we've laid out in the [previous section](#organize-the-big-picture-of-the-code), you may want to work on this iteratively as we dive into the code.
-5. As you clean up the code, you should run and re-run chunks to see if they work as you expect. You will also want to refresh your environment to help you develop the code (sometimes older objectives stuck in your environment can inhibit your ability to troubleshoot). In RStudio, you refresh your environment by going to the `Run` menu and using `Restart R and refresh clear output`.
+5. As you clean up the code, you should run and re-run chunks to see if they work as you expect. You will also want to refresh your environment to help you develop the code (sometimes older objects stuck in your environment can inhibit your ability to troubleshoot). In RStudio, you refresh your environment by going to the `Run` menu and using `Restart R and Clear Output`.
***
@@ -510,7 +510,7 @@ set.seed(1234)
**Get rid of setwd**
_Rationale:_
-`setwd()` almost never work for anyone besides the one person who wrote it. And in a few days/weeks it may not work for them either.
+`setwd()` almost never works for anyone besides the one person who wrote it. And in a few days/weeks it may not work for them either.
_Before:_
```
@@ -526,9 +526,7 @@ _Related readings:_
**Give the variables more informative names**
_Rationale:_
-`xx` doesn't tell us what is in the data here. Also by using the `readr::read_tsv()` from tidyverse we'll get a cleaner, faster read and won't have to specify `sep` argument. Note we are also fixing some spacing and using `<-` so that we can stick to readability conventions.
-
-You'll notice later
+`xx` doesn't tell us what is in the data here. Also by using the `readr::read_tsv()` from tidyverse we'll get a cleaner, faster read and won't have to specify the `sep` argument. Note we are also fixing some spacing and using `<-` so that we can stick to readability conventions.
_Before:_
```
@@ -551,10 +549,10 @@ What is happening with df1 and df2? What's being filtered out? etc.
Code comments would certainly help understanding, but even better, we can DRY this code up and make the code clearer on its own.
_Before:_
-It may be difficult to tell from looking at the before code because there are no comments and it's a bit tricky to read, but the goal of this is to:
+It may be difficult to tell from looking at the before code because there are no comments and it's a bit tricky to read, but the goal of this code is to:
-1) Calculate variances for each row (each row is a gene).
-2) Filter the original gene expression matrix to only genes have a bigger variance (here we use arbitrarily 10 as a filter cutoff).
+1) Calculate the variance for each row (each row is a gene with expression values from a number of samples).
+2) Filter the original gene expression matrix to only genes that have a bigger variance (here we arbitrarily use 10 as a filter cutoff).
```
df=read.csv("SRP070849.tsv", sep="\t")
@@ -575,10 +573,10 @@ Let's see how we can do this in a DRY'er and clearer way.
We can:
1) Add comments to describe our goals.
2) Use variable names that are more informative.
-3) Use the apply functions to do the loop for us -- this will eliminate the need for unclear variable `i` as well.
+3) Use the apply functions to do the loop for us -- this will eliminate the need for the unclear variable `i` as well.
4) Use the tidyverse to do the filtering for us so we don't have to rename data frames or store extra versions of `df`.
-Here's what the above might look like after some refactoring. Hopefully you find this is easier to follow and total there's less lines of code (but also has comments too!).
+Here's what the above might look like after some refactoring. Hopefully you find this is easier to follow and there's less total lines of code (but it also has comments now too!).
```
# Read in data TSV file
expression_df <- readr::read_tsv(data_file) %>%
diff --git a/08-readmes.Rmd b/08-readmes.Rmd
index 8d645a5..b5a47ae 100644
--- a/08-readmes.Rmd
+++ b/08-readmes.Rmd
@@ -15,11 +15,11 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
Documentation is an important but sometimes overlooked part of creating a reproducible analysis! There are two parts of documentation we will discuss here: 1) In notebook descriptions and 2) READMEs
-Both these notebook descriptions and READMEs are written in markdown -- a shorthand for html (the same as the documentation parts of your code). If you aren't familiar, markdown is such a handy tool and we encourage you to learn it (it doesn't take too long), here's a [quick guide](https://www.markdownguide.org/cheat-sheet/) to get you started.
+Both these notebook descriptions and READMEs are written in markdown -- a lightweight markup language for formatting text that can be converted to html for display in web browsers. If you aren't familiar, markdown is such a handy tool and we encourage you to learn it (it doesn't take too long), here's a [quick guide](https://www.markdownguide.org/cheat-sheet/) to get you started.
### Notebook descriptions
-As we [discussed in chapter 5](https://jhudatascience.org/Reproducibility_in_Cancer_Informatics/using-notebooks.html#reason-1-notebooks-allow-for-tracking-data-exploration-and-encourage-the-scientist-to-narrate-their-thought-process), data analyses can lead one on a winding trail of decisions, but notebooks allow you to narrate your thought process as you travel along these analyses explorations!
+As we [discussed in chapter 5](https://jhudatascience.org/Reproducibility_in_Cancer_Informatics/using-notebooks.html#reason-1-notebooks-allow-for-tracking-data-exploration-and-encourage-the-scientist-to-narrate-their-thought-process), data analyses can lead one on a winding trail of decisions, but notebooks allow you to narrate your thought process as you travel along these analysis explorations!
```{r, fig.align='center', echo = FALSE, fig.alt= "Ruby is looking at her computer that has a lovely notebook with a heatmap! Ruby says ‘Working from this notebook allows me to interactively develop on my data analysis and write down my thoughts about the process all in one place!’"}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf8f405fdab_0_186")
@@ -35,13 +35,13 @@ What scientific question are you trying to answer? Describe the dataset you are
Describe why a particular code chunk is doing a particular thing -- the more odd the code looks, the greater need for you to describe why you are doing it.
-Describe any particular filters or cutoffs you are using and how did you decide on those?
+Describe any particular filters or cutoffs you are using and how you decided on those?
For data wrangling steps, why are you wrangling the data in such a way -- is this because a certain package you are using requires it?
#### Your observations of the results
-What do you think about the results? The plots and tables you show in the notebook -- how do they inform your original questions?
+What do you think about the results? For the plots and tables you show in the notebook -- how do they inform your original questions?
### READMEs!
@@ -51,17 +51,27 @@ READMEs are also a great way to help your collaborators get quickly acquainted w
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf8379bb805_0_11")
```
-READMEs stick out in a project and are generally universal signal for new people to the project to start by READing them. GitHub automatically will preview your file called "README.md" when someone comes to the main page of your repository which further encourages people looking at your project to read the information in your README.
+READMEs stick out in a project and are generally universal signal for new people to the project to start by READing them. GitHub automatically will preview your file called "README.md" when someone comes to the main page of your repository, which further encourages people looking at your project to read the information in your README.
**Information that should be included in a README:**
1) General purpose of the project
2) Instructions on how to re-run the project
3) Lists of any software required by the project
-4) Input and output file descriptions.
-5) Descriptions of any additional tools included in the project?
+4) Input and output file descriptions
+5) Descriptions of any additional tools included in the project
-You can take a look at this [template README](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/resources/README-template.md) to get your started.
+You can take a look at this [template README](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/resources/README-template.md) to get started.
+
+## Licensing
+
+Adding information about a license is a good idea. If you put your code on GitHub, then the default copyright laws apply. According to GitHub:
+
+> “You retain all rights to your source code and no one may reproduce, distribute, or create derivative works from your work. If you’re creating an open source project, we strongly encourage you to include an open source license.”
+
+Open source software or code means that it is distributed with a license that allows others to reuse or adapt your code for other purposes. This is very helpful to advance science and technology.
+
+Check out this great resource on [options for licenses](https://choosealicense.com/) to help you choose which license is right for your project.
#### More about writing READMEs:
@@ -69,7 +79,7 @@ You can take a look at this [template README](https://raw.githubusercontent.com/
- [A Beginners Guide to writing a Kicka** README](https://meakaakka.medium.com/a-beginners-guide-to-writing-a-kickass-readme-7ac01da88ab3)
- [Tips for Creating Great README Docs](https://www.archbee.com/blog/readme-creating-tips)
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## Get the exercise project files
**Get the Python project example files**
[Click this link to download](https://raw.githubusercontent.com/jhudsl/Reproducibility_in_Cancer_Informatics/main/chapter-zips/python-heatmap-chapt-8.zip).
@@ -113,7 +123,7 @@ unzip -o chapter-zips/r-heatmap-chapt-8.zip -d chapter-zips/
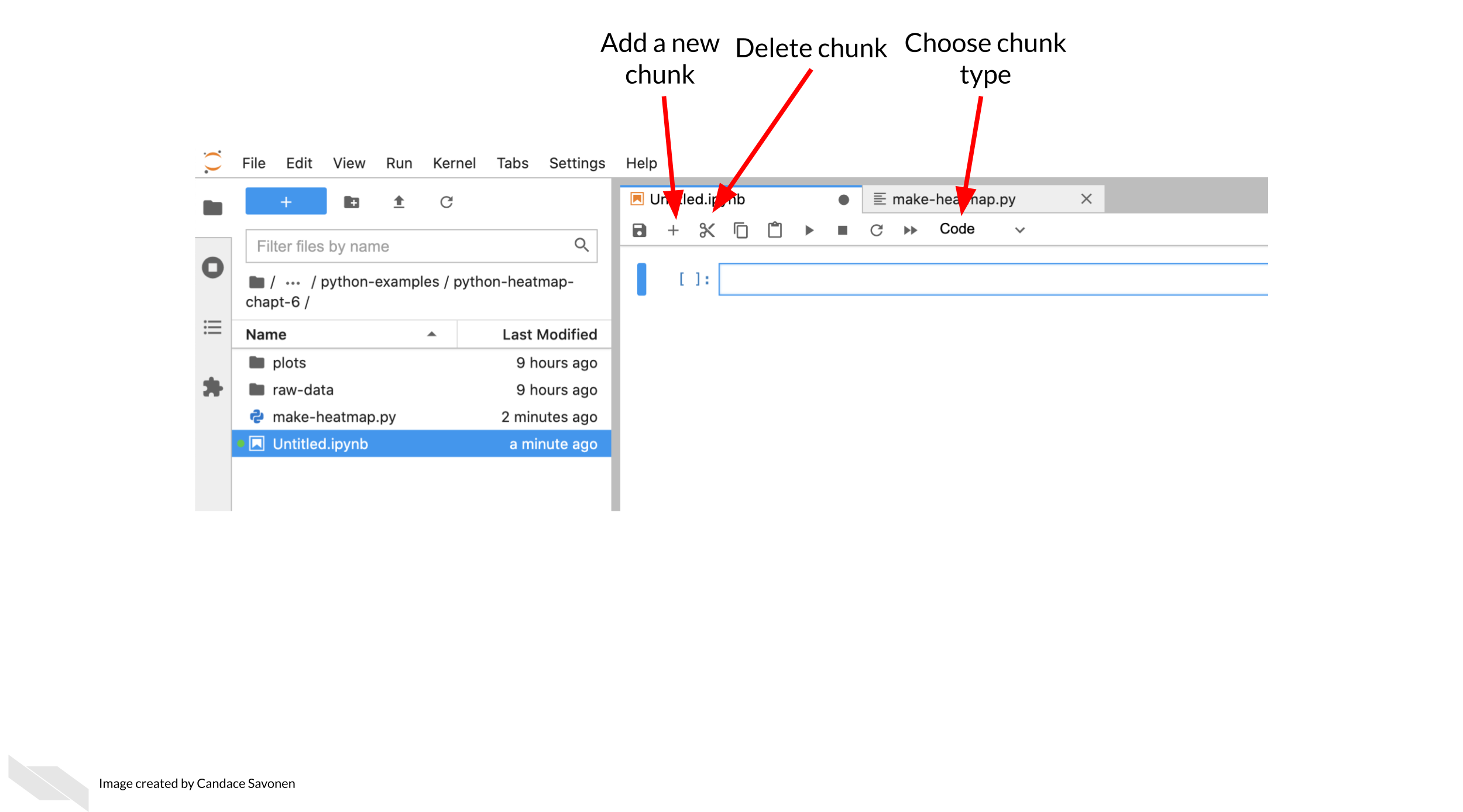
```{r, fig.align='center', fig.alt= "The Jupyter interface has a ‘add a new chunk’ button, a delete chunk button, and a dropdown menu that allows you to choose the chunk type you’d like to add. "}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gfaa026a583_0_30")
```
-5. Continue to add more descriptions where you feel is necessary, You can reference the descriptions we have in the ["final" version](https://github.com/jhudsl/reproducible-python-example/blob/main/make_heatmap.ipynb) looks like in the [example Python repository](https://github.com/jhudsl/reproducible-python-example). (Again, _final_ here is in quotes because we may continue to make improvements to this notebook too -- remember what we said about iterative?)
+5. Continue to add more descriptions where you feel is necessary. You can reference the descriptions we have in the ["final" version](https://github.com/jhudsl/reproducible-python-example/blob/main/make_heatmap.ipynb) from the [example Python repository](https://github.com/jhudsl/reproducible-python-example). (Again, _final_ here is in quotes because we may continue to make improvements to this notebook -- remember what we said about iterative?)
@@ -123,7 +133,7 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
2. Open up your notebook you've been working on in the previous chapters: `make_heatmap.Rmd`
3. In between code chunks, add more descriptions using Markdown language.
4. You can test how this renders by saving your `.Rmd` and then opening up the resulting `nb.html` file and choosing `View in Browser`.
-5. Continue to add more descriptions where you feel is necessary. You can reference the descriptions we have in the ["final" version](https://jhudatascience.org/reproducible-R-example/01-heatmap.nb.html) looks like in the [example R repository](https://github.com/jhudsl/reproducible-R-example). (Again, _final_ here is in quotes because we may continue to make improvements to this notebook too -- remember what we said about iterative?)
+5. Continue to add more descriptions where you feel is necessary. You can reference the descriptions we have in the ["final" version](https://jhudatascience.org/reproducible-R-example/01-heatmap.nb.html) from the [example R repository](https://github.com/jhudsl/reproducible-R-example). (Again, _final_ here is in quotes because we may continue to make improvements to this notebook -- remember what we said about iterative?)
diff --git a/09-code-review.Rmd b/09-code-review.Rmd
index 469a9ff..dcae603 100644
--- a/09-code-review.Rmd
+++ b/09-code-review.Rmd
@@ -11,7 +11,7 @@ ottrpal::set_knitr_image_path()
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.gf8379bb805_0_6")
```
-We've previously discussed that the only way to know if your analysis is truly reproducible is to send it to someone else to reproduce! That sentiment is at the heart of code review. Although most wouldn't dare send out a manuscript for publishing without having our collaborators giving it a line-by-line review, people don't always feel the same way about code.
+We've previously discussed that the only way to know if your analysis is truly reproducible is to send it to someone else to reproduce! That sentiment is at the heart of code review. Although most wouldn't dare send out a manuscript for publishing without having collaborators giving it a line-by-line review, people don't always feel the same way about code.
@Parker2017 describes code review:
@@ -34,7 +34,7 @@ Since reproducibility is all about someone else being able to run your code and
```{r, fig.align='center', echo = FALSE, fig.alt= "Ruby the researcher and Avi the associate are so happy because Avi was able to reproduce Ruby’s results on his computer."}
ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF6KS9QmJ5NBjwWVjRn40MS9f2noE/edit#slide=id.g1006ff8e7e9_48_33")
```
-In the second part of this reproducibility course we will discuss how to conduct formal line-by-line code reviews, but for now, we will discuss how to prep your analysis for someone else look at your code and attempt to run it.
+In the second part of this reproducibility course we will discuss how to conduct formal line-by-line code reviews, but for now, we will discuss how to prep your analysis for someone else to look at your code and attempt to run it.
At this point, you should have a GitHub repository that contains the following:
@@ -49,19 +49,19 @@ Before you send off your code to someone else, delete your output (the `results`
**2) Re-run the whole analysis**
-**3) Interrogate and troubleshoot**
+**3) Interrogate and troubleshoot**
If your code has any issues running, try your best to troubleshoot the problems. Read this handy guide for [tips on troubleshooting R](https://github.com/AlexsLemonade/training-modules/blob/master/intro-to-R-tidyverse/00b-debugging_resources.Rmd).
**4) Rinse and repeat**
-Repeat this as many times as needed until you reliably are able to re-run this code and get the same results without any [code smells](https://github.com/jennybc/code-smells-and-feels#code-smells-and-feels) popping up. Dig into bad code smells or bad results smells wherever you sense them. If you aren't sure why you feel this way about your code or results, hold on to this and it may be something your collaborator will be able to see something you don't.
+Repeat this as many times as needed until you reliably are able to re-run this code and get the same results without any [code smells](https://github.com/jennybc/code-smells-and-feels#code-smells-and-feels) popping up. Dig into bad code smells or bad results smells wherever you sense them. If you aren't sure why you feel this way about your code or results, hold on to this. It may be something your collaborator will be able to see that you missed.
**5) Let it simmer**
Leave your analysis for a bit. Do you think it's perfect? Are you at your wits end with it? No matter how you feel about it, let it sit for a half a day or so then return to it with fresh eyes [@Savonen2021b].
-**5) Re-review your documentation and code with fresh eyes**
+**6) Re-review your documentation and code with fresh eyes**
Now with fresh eyes and doing your best to imagine you don't have the knowledge you have -- does your analysis and results make sense?
-**6) Are you sure it's ready?**
+**7) Are you sure it's ready?**
Ask yourself if you've polished this code and documentation as far as you can reasonably take it? Realizing that determining what qualifies `as far as you can reasonably take it` is also a skill you will build with time. Code review is the most efficient use of everyone's time when your code and documentation have reached this point.
**8) Draft your request**
@@ -79,7 +79,7 @@ _Code review requests should include:_
- Lastly, thank them for helping review your code!
**9) Ready for review**
-Now you are ready to send your crafted message to your collaborator for review. But, for the purposes of this exercise, you may **not** want to ask your collaborator to spend their time carefully review this practice repository, but now that you understand and have done the steps involved _you are prepared to do this for your own analyses_!
+Now you are ready to send your crafted message to your collaborator for review. For the purposes of this exercise, you may **not** want to ask your collaborator to spend their time carefully reviewing a practice repository. However, now that you understand and have done the steps involved _you are prepared to do this for your own analyses_!
TL;DR for asking for a code review:
```{r, fig.align='center', echo = FALSE, fig.alt= "This chapter will demonstrate how to: How code review is the best way to enhance the reproducibility of your analyses over time. Engage in basic code review for your analysis."}
diff --git a/chapter-zips/python-heatmap-chapt-3.zip b/chapter-zips/python-heatmap-chapt-3.zip
index e41d67f..249d7d0 100644
Binary files a/chapter-zips/python-heatmap-chapt-3.zip and b/chapter-zips/python-heatmap-chapt-3.zip differ
diff --git a/chapter-zips/r-heatmap-chapt-3.zip b/chapter-zips/r-heatmap-chapt-3.zip
index 2232d62..ca94354 100644
Binary files a/chapter-zips/r-heatmap-chapt-3.zip and b/chapter-zips/r-heatmap-chapt-3.zip differ
 Getting your code to work the first time is the first step, but don't stop there!
-Just like in writing a manuscript you wouldn't consider your first draft a final draft, your polishing code works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
+Just like in writing a manuscript you wouldn't consider your first draft a final draft, your code polishing works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
#### Prioritize readability over cleverness
Getting your code to work the first time is the first step, but don't stop there!
-Just like in writing a manuscript you wouldn't consider your first draft a final draft, your polishing code works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
+Just like in writing a manuscript you wouldn't consider your first draft a final draft, your code polishing works best in an iterative manner. Although you may need to set it aside for the day to give your brain a rest, return to your code later with fresh eyes and try to look for ways to improve upon it!
#### Prioritize readability over cleverness
 -Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness comprises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, including your future self!).
+Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness compromises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, this includes your future self!).
What does readable code look like? @Orosz2019 has some thoughts on writing readable code:
@@ -34,7 +34,7 @@ What does readable code look like? @Orosz2019 has some thoughts on writing reada
> **The real test of readable code is others reading it.** So get feedback from others, via code reviews. Ask people to share feedback on how clear the code is. Encourage people to ask questions if something does not make sense. Code reviews - especially thorough code reviews - are the best way to get feedback on how good and readable your code is.
>
-> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints to opportunities to make the code more readable.
+> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints at opportunities to make the code more readable.
>
> A good way to get more feedback on the clarity of your code is to ask for feedback from someone who is not an expert on the codebase you are working on. Ask specifically for feedback on how easy to read your code is. Because this developer is not an expert on the codebase, they'll focus on how much they can follow your code. Most of the comments they make will be about your code's readability.
@@ -60,7 +60,7 @@ If you find yourself writing something more than once, you might want to write a
DRY code is easier on the reviewer because they don't have to review the same thing twice, but also because they don't have to review the same thing twice. ;)
DRYing code is something that takes some iterative passes and edits through your code, but in the end DRY code saves you and your collaborators time and can be something you reuse again in a future project!
-Here's an slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
+Here's a slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
```
paste('Hello','John', 'welcome to this course')
@@ -109,9 +109,9 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
-Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness comprises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, including your future self!).
+Some cleverness in code can be helpful, too much can make it difficult for others (including your future self!) to understand. If cleverness compromises the readability of your code, it probably is not worth it. Clever but unreadable code won't be re-used or trusted by others (AGAIN, this includes your future self!).
What does readable code look like? @Orosz2019 has some thoughts on writing readable code:
@@ -34,7 +34,7 @@ What does readable code look like? @Orosz2019 has some thoughts on writing reada
> **The real test of readable code is others reading it.** So get feedback from others, via code reviews. Ask people to share feedback on how clear the code is. Encourage people to ask questions if something does not make sense. Code reviews - especially thorough code reviews - are the best way to get feedback on how good and readable your code is.
>
-> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints to opportunities to make the code more readable.
+> Readable code will attract little to no clarifying questions, and reviewers won't misunderstand it. So pay careful attention to the cases when you realize someone misunderstood the intent of what you wrote or asked a clarifying question. Every question or misunderstanding hints at opportunities to make the code more readable.
>
> A good way to get more feedback on the clarity of your code is to ask for feedback from someone who is not an expert on the codebase you are working on. Ask specifically for feedback on how easy to read your code is. Because this developer is not an expert on the codebase, they'll focus on how much they can follow your code. Most of the comments they make will be about your code's readability.
@@ -60,7 +60,7 @@ If you find yourself writing something more than once, you might want to write a
DRY code is easier on the reviewer because they don't have to review the same thing twice, but also because they don't have to review the same thing twice. ;)
DRYing code is something that takes some iterative passes and edits through your code, but in the end DRY code saves you and your collaborators time and can be something you reuse again in a future project!
-Here's an slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
+Here's a slightly modified example from @Bernardo2021 for what DRY vs non-DRY code might look like:
```
paste('Hello','John', 'welcome to this course')
@@ -109,9 +109,9 @@ ottrpal::include_slide("https://docs.google.com/presentation/d/1LMurysUhCjZb7DVF
 -Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may even work all the same, just like you understood what I wrote in that last sentence, but a lack of consistent style can make require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
+Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may run correctly, just like you understood what I wrote in that last sentence, but a lack of consistent style can require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
-There are different style guides out there that people adhere to. It doesn't matter so much which one you choose, so much that you pick one and stick to it for a particular project.
+There are different style guides out there that people adhere to. It doesn't matter which one you choose, as long as you pick one and stick to it for a particular project.
_Python style guides_:
@@ -212,7 +212,7 @@ _R style guides_:
- [Hadley Wickham's Style guide](http://adv-r.had.co.nz/Style.html) @Wickham.
- [Google R style guide](https://google.github.io/styleguide/Rguide.html) @GoogleR.
-Although writing code following a style as you are writing is a good practice, we're all human and that can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
+Although writing code that follows a style is a good practice, we're all human and it can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
For Python code, you can use [python black](https://black.readthedocs.io/en/stable/) and for R, [styler](https://www.tidyverse.org/blog/2017/12/styler-1.0.0/).
#### Organize the structure of your code
@@ -304,7 +304,7 @@ There's so many opinions and strategies on best practices for code. And although
- [R for Epidemiology: Coding best practices](https://www.r4epi.com/chapters/best_practices/best_practices) by @Cannell2021.
- [Best practices for R Programming](https://towardsdatascience.com/best-practices-for-r-programming-ec0754010b5a) by @Bernardo2021.
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## See example project files
-Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may even work all the same, just like you understood what I wrote in that last sentence, but a lack of consistent style can make require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
+Just like when writing doesN"t FoLLOW conv3nTi0Ns OR_sPAcinng 0r sp3llinG, it can be distracting, the same goes for code. Your code may run correctly, just like you understood what I wrote in that last sentence, but a lack of consistent style can require more brain power from your readers for them to understand. For reproducibility purposes, readability is important! The easier you can make it on your readers, the more likely they will be able to understand and reproduce the results.
-There are different style guides out there that people adhere to. It doesn't matter so much which one you choose, so much that you pick one and stick to it for a particular project.
+There are different style guides out there that people adhere to. It doesn't matter which one you choose, as long as you pick one and stick to it for a particular project.
_Python style guides_:
@@ -212,7 +212,7 @@ _R style guides_:
- [Hadley Wickham's Style guide](http://adv-r.had.co.nz/Style.html) @Wickham.
- [Google R style guide](https://google.github.io/styleguide/Rguide.html) @GoogleR.
-Although writing code following a style as you are writing is a good practice, we're all human and that can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
+Although writing code that follows a style is a good practice, we're all human and it can be tricky to do, so we recommend using an automatic styler on your code to fix up your code for you.
For Python code, you can use [python black](https://black.readthedocs.io/en/stable/) and for R, [styler](https://www.tidyverse.org/blog/2017/12/styler-1.0.0/).
#### Organize the structure of your code
@@ -304,7 +304,7 @@ There's so many opinions and strategies on best practices for code. And although
- [R for Epidemiology: Coding best practices](https://www.r4epi.com/chapters/best_practices/best_practices) by @Cannell2021.
- [Best practices for R Programming](https://towardsdatascience.com/best-practices-for-r-programming-ec0754010b5a) by @Bernardo2021.
-## Get the exercise project files (or continue with the files you used in the previous chapter)
+## See example project files