diff --git a/.github/CONTRIBUTING.md b/.github/CONTRIBUTING.md
index 8fe24ff..f7e82ce 100644
--- a/.github/CONTRIBUTING.md
+++ b/.github/CONTRIBUTING.md
@@ -1,10 +1,9 @@
# Contribution Guidelines
**All people are very much welcome to contribute to code, documentation, testing and suggestions.**
-
+`psycho` is a beginner-friendly package. Even if you're new to all this open-source way of life, new to coding and submitting pull requests (PRs), we encourage you to try. You can submit some recommendations in the [issues](https://github.com/neuropsychology/psycho.R/issues) or start coding a function or adding some parameters, fixing a bug or just rewriting a bit so it is more clear, more documented or understandable. Then, do not hesitate to sumbit your changes. We will work together to integrate them :)
## Code
- Authors of code contribution will be added within the [**authors**](https://github.com/neuropsychology/psycho.R/blob/master/DESCRIPTION) section within the package description.
- Please document and comment your code, so that the purpose of each step (or code line) is stated in a clear and understandable way.
-- Avoid unnecessary function splitting into smaller bits: it makes testing and reading of the code more difficult (as one must jump between functions and files to understand what's happening).
- Before submitting a change, please use [**styler**](https://github.com/r-lib/styler) to nicely format your code.
\ No newline at end of file
diff --git a/docs/_posts/2018-06-11-bayesian_correlation.md b/docs/_posts/2018-06-11-bayesian_correlation.md
index 98217be..ea58b7a 100644
--- a/docs/_posts/2018-06-11-bayesian_correlation.md
+++ b/docs/_posts/2018-06-11-bayesian_correlation.md
@@ -92,6 +92,13 @@ plot(density(posterior))
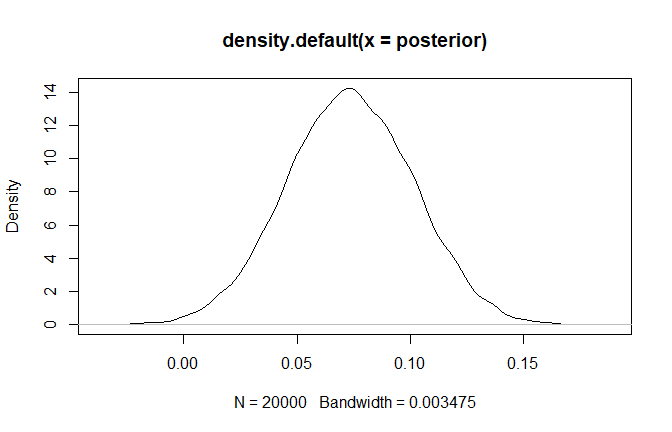 +
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues) or, even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
Credits
=======
diff --git a/docs/_posts/preparation/analyze_correlation.Rmd b/docs/_posts/preparation/analyze_correlation.Rmd
index 9fb10b0..cb8767a 100644
--- a/docs/_posts/preparation/analyze_correlation.Rmd
+++ b/docs/_posts/preparation/analyze_correlation.Rmd
@@ -2,12 +2,12 @@
title: "APA Formatted Correlation"
layout: post
output:
- html_document:
- df_print: paged
- toc: yes
md_document:
toc: yes
variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
author: "Dominique Makowski"
date: "`r Sys.Date()`"
editor_options:
@@ -57,6 +57,9 @@ knitr::kable(summary(results), digits=2)
Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
# Credits
diff --git a/docs/_posts/preparation/analyze_correlation.knit.md b/docs/_posts/preparation/analyze_correlation.knit.md
new file mode 100644
index 0000000..d61145f
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.knit.md
@@ -0,0 +1,77 @@
+---
+title: "APA Formatted Correlation"
+layout: post
+output:
+ md_document:
+ toc: yes
+ variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
+author: "Dominique Makowski"
+date: "2018-06-11"
+editor_options:
+ chunk_output_type: console
+---
+
+
+
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+# Do a correlation
+
+```r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+
+# APA formatted output
+
+
+```r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+```
+The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+```
+
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+# Dataframe of Values
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+
+```r
+results <- analyze(cor_results)
+summary(results)
+```
+
+ effect statistic df p CI_lower CI_higher
+------- ---------- ---- --- --------- ----------
+ 0.87 21.65 148 0 0.83 0.91
+
+# Bayesian Correlation
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
+# Credits
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470. https://doi.org/10.21105/joss.00470
diff --git a/docs/_posts/preparation/analyze_correlation.md b/docs/_posts/preparation/analyze_correlation.md
new file mode 100644
index 0000000..aafebd6
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.md
@@ -0,0 +1,64 @@
+- [Do a correlation](#do-a-correlation)
+- [APA formatted output](#apa-formatted-output)
+- [Dataframe of Values](#dataframe-of-values)
+- [Bayesian Correlation](#bayesian-correlation)
+- [Contribute](#contribute)
+- [Credits](#credits)
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+Do a correlation
+================
+
+``` r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+APA formatted output
+====================
+
+``` r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+ The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+Dataframe of Values
+===================
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+``` r
+results <- analyze(cor_results)
+summary(results)
+```
+
+| effect| statistic| df| p| CI\_lower| CI\_higher|
+|-------:|----------:|----:|----:|----------:|-----------:|
+| 0.87| 21.65| 148| 0| 0.83| 0.91|
+
+Bayesian Correlation
+====================
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+Credits
+=======
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470.
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues) or, even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
Credits
=======
diff --git a/docs/_posts/preparation/analyze_correlation.Rmd b/docs/_posts/preparation/analyze_correlation.Rmd
index 9fb10b0..cb8767a 100644
--- a/docs/_posts/preparation/analyze_correlation.Rmd
+++ b/docs/_posts/preparation/analyze_correlation.Rmd
@@ -2,12 +2,12 @@
title: "APA Formatted Correlation"
layout: post
output:
- html_document:
- df_print: paged
- toc: yes
md_document:
toc: yes
variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
author: "Dominique Makowski"
date: "`r Sys.Date()`"
editor_options:
@@ -57,6 +57,9 @@ knitr::kable(summary(results), digits=2)
Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
# Credits
diff --git a/docs/_posts/preparation/analyze_correlation.knit.md b/docs/_posts/preparation/analyze_correlation.knit.md
new file mode 100644
index 0000000..d61145f
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.knit.md
@@ -0,0 +1,77 @@
+---
+title: "APA Formatted Correlation"
+layout: post
+output:
+ md_document:
+ toc: yes
+ variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
+author: "Dominique Makowski"
+date: "2018-06-11"
+editor_options:
+ chunk_output_type: console
+---
+
+
+
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+# Do a correlation
+
+```r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+
+# APA formatted output
+
+
+```r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+```
+The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+```
+
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+# Dataframe of Values
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+
+```r
+results <- analyze(cor_results)
+summary(results)
+```
+
+ effect statistic df p CI_lower CI_higher
+------- ---------- ---- --- --------- ----------
+ 0.87 21.65 148 0 0.83 0.91
+
+# Bayesian Correlation
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
+# Credits
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470. https://doi.org/10.21105/joss.00470
diff --git a/docs/_posts/preparation/analyze_correlation.md b/docs/_posts/preparation/analyze_correlation.md
new file mode 100644
index 0000000..aafebd6
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.md
@@ -0,0 +1,64 @@
+- [Do a correlation](#do-a-correlation)
+- [APA formatted output](#apa-formatted-output)
+- [Dataframe of Values](#dataframe-of-values)
+- [Bayesian Correlation](#bayesian-correlation)
+- [Contribute](#contribute)
+- [Credits](#credits)
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+Do a correlation
+================
+
+``` r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+APA formatted output
+====================
+
+``` r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+ The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+Dataframe of Values
+===================
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+``` r
+results <- analyze(cor_results)
+summary(results)
+```
+
+| effect| statistic| df| p| CI\_lower| CI\_higher|
+|-------:|----------:|----:|----:|----------:|-----------:|
+| 0.87| 21.65| 148| 0| 0.83| 0.91|
+
+Bayesian Correlation
+====================
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+Credits
+=======
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470.
 +
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues) or, even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
Credits
=======
diff --git a/docs/_posts/preparation/analyze_correlation.Rmd b/docs/_posts/preparation/analyze_correlation.Rmd
index 9fb10b0..cb8767a 100644
--- a/docs/_posts/preparation/analyze_correlation.Rmd
+++ b/docs/_posts/preparation/analyze_correlation.Rmd
@@ -2,12 +2,12 @@
title: "APA Formatted Correlation"
layout: post
output:
- html_document:
- df_print: paged
- toc: yes
md_document:
toc: yes
variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
author: "Dominique Makowski"
date: "`r Sys.Date()`"
editor_options:
@@ -57,6 +57,9 @@ knitr::kable(summary(results), digits=2)
Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
# Credits
diff --git a/docs/_posts/preparation/analyze_correlation.knit.md b/docs/_posts/preparation/analyze_correlation.knit.md
new file mode 100644
index 0000000..d61145f
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.knit.md
@@ -0,0 +1,77 @@
+---
+title: "APA Formatted Correlation"
+layout: post
+output:
+ md_document:
+ toc: yes
+ variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
+author: "Dominique Makowski"
+date: "2018-06-11"
+editor_options:
+ chunk_output_type: console
+---
+
+
+
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+# Do a correlation
+
+```r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+
+# APA formatted output
+
+
+```r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+```
+The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+```
+
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+# Dataframe of Values
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+
+```r
+results <- analyze(cor_results)
+summary(results)
+```
+
+ effect statistic df p CI_lower CI_higher
+------- ---------- ---- --- --------- ----------
+ 0.87 21.65 148 0 0.83 0.91
+
+# Bayesian Correlation
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
+# Credits
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470. https://doi.org/10.21105/joss.00470
diff --git a/docs/_posts/preparation/analyze_correlation.md b/docs/_posts/preparation/analyze_correlation.md
new file mode 100644
index 0000000..aafebd6
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.md
@@ -0,0 +1,64 @@
+- [Do a correlation](#do-a-correlation)
+- [APA formatted output](#apa-formatted-output)
+- [Dataframe of Values](#dataframe-of-values)
+- [Bayesian Correlation](#bayesian-correlation)
+- [Contribute](#contribute)
+- [Credits](#credits)
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+Do a correlation
+================
+
+``` r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+APA formatted output
+====================
+
+``` r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+ The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+Dataframe of Values
+===================
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+``` r
+results <- analyze(cor_results)
+summary(results)
+```
+
+| effect| statistic| df| p| CI\_lower| CI\_higher|
+|-------:|----------:|----:|----:|----------:|-----------:|
+| 0.87| 21.65| 148| 0| 0.83| 0.91|
+
+Bayesian Correlation
+====================
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+Credits
+=======
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470.
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues) or, even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
Credits
=======
diff --git a/docs/_posts/preparation/analyze_correlation.Rmd b/docs/_posts/preparation/analyze_correlation.Rmd
index 9fb10b0..cb8767a 100644
--- a/docs/_posts/preparation/analyze_correlation.Rmd
+++ b/docs/_posts/preparation/analyze_correlation.Rmd
@@ -2,12 +2,12 @@
title: "APA Formatted Correlation"
layout: post
output:
- html_document:
- df_print: paged
- toc: yes
md_document:
toc: yes
variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
author: "Dominique Makowski"
date: "`r Sys.Date()`"
editor_options:
@@ -57,6 +57,9 @@ knitr::kable(summary(results), digits=2)
Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
# Credits
diff --git a/docs/_posts/preparation/analyze_correlation.knit.md b/docs/_posts/preparation/analyze_correlation.knit.md
new file mode 100644
index 0000000..d61145f
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.knit.md
@@ -0,0 +1,77 @@
+---
+title: "APA Formatted Correlation"
+layout: post
+output:
+ md_document:
+ toc: yes
+ variant: markdown_github
+ html_document:
+ df_print: paged
+ toc: yes
+author: "Dominique Makowski"
+date: "2018-06-11"
+editor_options:
+ chunk_output_type: console
+---
+
+
+
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+# Do a correlation
+
+```r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+
+# APA formatted output
+
+
+```r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+```
+The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+```
+
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+# Dataframe of Values
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+
+```r
+results <- analyze(cor_results)
+summary(results)
+```
+
+ effect statistic df p CI_lower CI_higher
+------- ---------- ---- --- --------- ----------
+ 0.87 21.65 148 0 0.83 0.91
+
+# Bayesian Correlation
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+# Contribute
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+
+# Credits
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470. https://doi.org/10.21105/joss.00470
diff --git a/docs/_posts/preparation/analyze_correlation.md b/docs/_posts/preparation/analyze_correlation.md
new file mode 100644
index 0000000..aafebd6
--- /dev/null
+++ b/docs/_posts/preparation/analyze_correlation.md
@@ -0,0 +1,64 @@
+- [Do a correlation](#do-a-correlation)
+- [APA formatted output](#apa-formatted-output)
+- [Dataframe of Values](#dataframe-of-values)
+- [Bayesian Correlation](#bayesian-correlation)
+- [Contribute](#contribute)
+- [Credits](#credits)
+
+One of the most time-consuming part of data analysis in psychology is the copy-pasting of specific values of some R output to a manuscript or a report. This task is frustrating, prone to errors, and increase de variability of statistical reporting. At the sime time, standardizing practices of what and how to report is crucial for reproducibility and clarity. **The [psycho](https://github.com/neuropsychology/psycho.R) package was designed specifically to do this job**, at first for complex [Bayesian mixed models](https://cran.r-project.org/web/packages/psycho/vignettes/bayesian.html), but is now also compatible with basic methods, such as **correlation**.
+
+Do a correlation
+================
+
+``` r
+df <- iris
+cor_results <- cor.test(df$Sepal.Length, df$Petal.Length)
+```
+
+APA formatted output
+====================
+
+``` r
+# devtools::install_github("neuropsychology/psycho.R") # Install the latest psycho version
+
+# Load packages
+library(psycho)
+psycho::analyze(cor_results)
+```
+
+ The Pearson's product-moment correlation between df$Sepal.Length and df$Petal.Length is significantly large and positive (r(148) = 0.87, 95% CI [0.83, 0.91], p < .001)
+
+The formatted output includes **direction**, **effect size** (interpreted by default with **[Cohen's (1988)](https://github.com/neuropsychology/psycho.R/blob/master/R/interpret_r.R#L142)** rules) and **confidence intervals**. Now you can just copy and paste this line into your report and focus on what's important.
+
+Dataframe of Values
+===================
+
+It is also possible to have all the values stored in a dataframe by running a **summary** on the analyzed object.
+
+``` r
+results <- analyze(cor_results)
+summary(results)
+```
+
+| effect| statistic| df| p| CI\_lower| CI\_higher|
+|-------:|----------:|----:|----:|----------:|-----------:|
+| 0.87| 21.65| 148| 0| 0.83| 0.91|
+
+Bayesian Correlation
+====================
+
+Nevertheless, we recommand doing a **Bayesian correlation**, which is even [easier and quicker to do](blogpostlink)!
+
+Contribute
+==========
+
+Of course, these reporting standards should change, depending on new expert recommandations or official guidelines. **The goal of this package is to flexibly adaptive to new changes and good practices evolution**. Therefore, if you have any advices, opinions or such, we encourage you to either let us know by opening an [issue](https://github.com/neuropsychology/psycho.R/issues), or even better, try to implement them yourself by [contributing](https://github.com/neuropsychology/psycho.R/blob/master/.github/CONTRIBUTING.md) to the code.
+
+Credits
+=======
+
+This package helped you? Don't forget to cite the various packages you used :)
+
+You can cite `psycho` as follows:
+
+- Makowski, (2018). *The psycho Package: An Efficient and Publishing-Oriented Workflow for Psychological Science*. Journal of Open Source Software, 3(22), 470.