Description
Over on discourse, Argantonio65 reported a problem when trying out a toy coin-flipping example. In playing around with it a bit, it seems that a numpy array consisting exclusively of ones[zeros] is treated as a single (scalar) one[zero] regardless of the actual length of the numpy array. Adding even a single zero[one] seems to cause the true length to be acknowledged. Similarly, wrapping the array in a pm.Data object also seems to yield correct behavior.
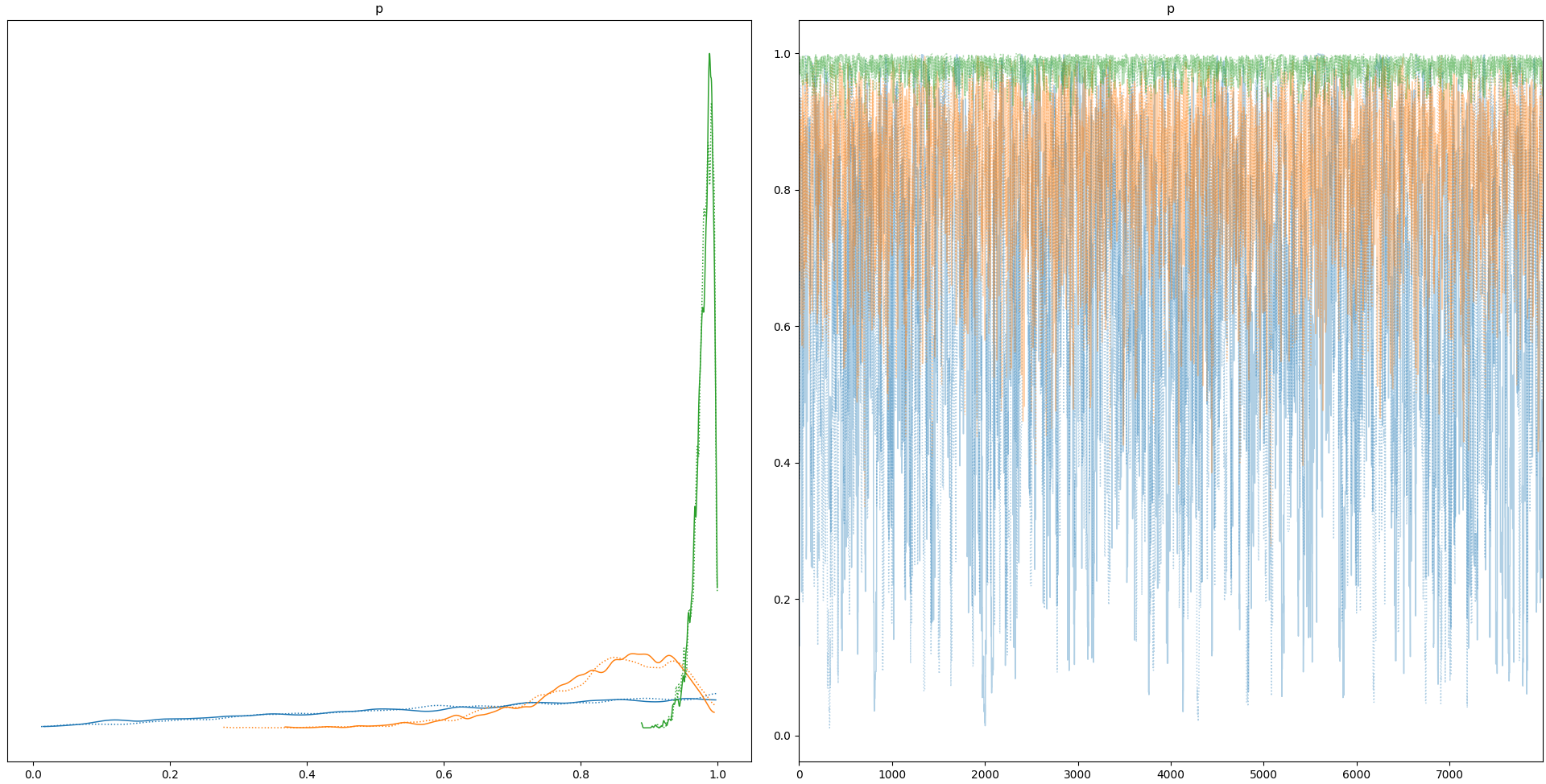
Here is a condensed version illustrating the insensitivity to sample size when the data is just a vector of ones.
import pymc3 as pm
import numpy as np
import matplotlib.pyplot as plt
data = [
np.ones(1),
np.ones(10),
np.ones(100)
]
with pm.Model() as model:
p = pm.Beta('p', alpha=1, beta=1, shape=3)
y1 = pm.Bernoulli('y1', p=p[0], observed=data[0])
y2 = pm.Bernoulli('y2', p=p[1], observed=data[1])
y3 = pm.Bernoulli('y3', p=p[2], observed=data[2])
trace = pm.sample(8000, step = pm.Metropolis(),random_seed=333, chains=2)
pm.traceplot(trace)
plt.show()This displays the following:
Changing the 3 likelihood lines so that they use pm.Data:
y1 = pm.Bernoulli('y1', p=p[0], observed=pm.Data('d1', data[0]))
y2 = pm.Bernoulli('y2', p=p[1], observed=pm.Data('d2', data[1]))
y3 = pm.Bernoulli('y3', p=p[2], observed=pm.Data('d3', data[2]))yields the expected behavior:
Augmenting the data with a single tail/failure:
data[1][0] = 0
data[2][0] = 0also yields the expected behavior.
Versions and main components
- PyMC3 Version:3.10
- Theano Version:1.0.5
- Theano-pymc Version:1.0.11
- Python Version:3.8.5
- Operating system:Linux
- How did you install PyMC3: pip