-
Notifications
You must be signed in to change notification settings - Fork 4
Installation guidelines
Some of you may need additional guidelines for installing Panache, here is a complement to what is already in the ReadMe.
Make sure you are using your IP in nginx.conf if you wish to make Panache available to other people. "localhost" can be used if you only need it on your personal machine.
In rare cases an Invalid Host header error can show up when running the container and trying to access the web page. This can be bypassed by adding disableHostCheck: true in a vue.config.js file at the root (which tells npm wich port and IP to use, 0.0.0.0 being localhost). See an example below:
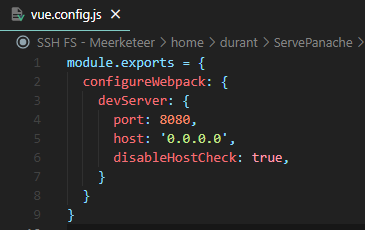
If you wish to use another web server directly with the production version of files instead, here are the steps to follow to create them:
npm install
npm install @vue/cli
npm run build #Creates the production version filesThe production files will be gathered into a new directory called dist, ready to be served.
You are likely to have problems with the paths to the different files in the newly created index.js, which may not have the right root for them. This is because the main path inferred by Vue by default may not match yours if it is in subfolders.
EDIT: This should be easily corrected by adding your desired root within the file vue.config.js, using publicPath. Check out Vue's docs for more details. Below is an exemple of vue.config.js:
module.exports = {
publicPath : "/test/panache/", // <-- The app will be served from the subfolder /test/panache/
configureWebpack: { // <-- General configuration properties for deployment
devServer: {
port: 8080,
host: '0.0.0.0',
disableHostCheck: true,
}
}
}Former, deprecated workaround:
You may for example see
/cssand/jsinstead ofcss/andjs/. You may also want to modify this line -->{path:"/ ",name:"Panache",component:xe["default"]}<-- injs/app.{randomID}.js, replacing the root with the directory containing the files to serve. For example, if the subpath hosting the server is called 'bananache' you should write{path:"/bananache/",name:"Panache",component:xe["default"]}instead. Kudos to Philipp Bayer for finding this one. We are looking for ways to correct this behaviour induced by npm.
If your goal is to set an instance of Panache that runs with only one set of files, you can use a dedicated version which does not offer the possibility to upload files but works from pre-formatted files instead. Our Banana Genome Hub version works this way, and is based on the alternative bananaGenomeHub branch. Note: as of 1st September 2021 the main branch is ahead of bananaGenomeHub, new changes will be cascaded soon.
Here you'll wish...
- To pre format your dataset into a json format
- To preset the store according to these json
Unfortunately there is no script to pre-format your dataset, yet. We propose two alternatives: copy json from the console, or directly load it as dedicated files (might work better for big files).
A conversion script is now available within the companionScript folder. Use vanillaFiles2JsonConverter.py with the files you would like to preformat into JSONs. It replaces existing solutions (see below) with a faster and safer method that you can run on your machine without depending on your browser's performance.
It can be used to produced a json file for your BED-like presence absence matrix, and the corresponding gff3 file. Standard output includes progress bars and list of CHROMOSOME_NAMES and GENOME_NAMES that are needed for the store.
The script is to be used with python3.6+, and uses the following parameters:
usage: vanillaFiles2JsonConverter.py pavName [-h] [--gffName GFFNAME] [--outPav OUTPAV] [--outGff OUTGFF]
positional arguments:
pavName BED-like file built for Panache that should be converted
into JSON format
optional arguments:
-h, --help show this help message and exit
--gffName GFFNAME If available, an additional gff file to convert alongside
the main pav file into JSON format
--outPav OUTPAV Output name for the formatted pav file. '.json' extension
will be added automatically. Default name is the same
than the input name.
--outGff OUTGFF Output name for the formatted gff file. '.json' extension
will be added automatically. Default name is the same
than the input name.A quick and dirty solution for this however is simply to run the default version of Panache, upload the desired files, and have them printed back as JSON once they are added to the store (ie. once they are fully loaded, parsed and formated into something usable by Panache). If you wish to try it at home I would advise to add the following to the default version:
... within PavFileParser.vue:
- Look for
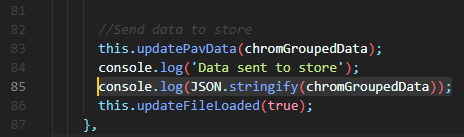
'parseDataToJson(loadedFile)', which is the method for the event doing the parsing and sending the new object to the store. - Right before or after
this.updatePavData(chromGroupedData)which will send the formated object to the store, you can extract the desired JSON - One way of doing so is by console.logging this object as a JSON with
console.log(JSON.stringify(chromGroupedData));:
... within GffFileParser.vue, similarly:
- Look for
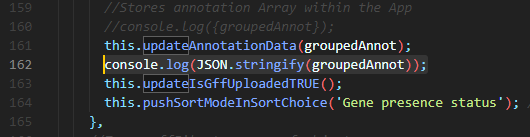
'parseGffToAnnotationObjects: async function(gffFile)' - Right before or after
this.updateAnnotationData(groupedAnnot)extract your JSON - You can write
console.log(JSON.stringify(groupedAnnot));:
... then run Panache, upload both default files within the web app (Pav file, then gff file by clicking the + button to show additional upload options), copy-paste the JSONs from the console into actual files, and voilà!
DISCLAIMER: This solution may not work for biggest files, below is an alternative "unholy solution" proposed by Philipp Bayer that would directly download the JSON without having to print to the console:
I put this piece of code into the
js/app.{randomID}.jsdirectly aroundthis.updatePavData(chromGroupedData)andthis.updateAnnotationData(groupedAnnot)as indicated:const jsonStr = JSON.stringify(c); const filename='data.json'; let element = document.createElement('a'); element.setAttribute('href', 'data:text/plain;charset=utf-8,' + encodeURIComponent(jsonStr)); element.setAttribute('download', filename); element.style.display = 'none'; document.body.appendChild(element); element.click(); document.body.removeChild(element);(or
JSON.stringify(u)for the gene annotation) That makes a fake link and downloads the data directly. [...] Right-click and copy from the console would cause my Firefox to crash with a 100Mb json file. As a reminder,js/app.{randomID}.jsis a file located in thedistfolder after runningnpm run build. The script might have to be one one line.
Using the bananaGenomeHub branch as a basis, you will have to modify LocalFilter.vue with the information from your dataset in order to override the data sent to store when the app is mounted. You can follow the example made for the Banana Genome Hub as shown below:
async updateBananaData() {
//Enable loadingSpinner in Panache view
this.updateDisplayLoadingStatus();
//Send parameters not directly extracted from the data files
this.updateChromNames([
'chr01', 'chr02', 'chr03', 'chr04', 'chr05', 'chr06', 'chr07', 'chr08',
'chr09', 'chr10', 'chr11', 'chrUn_random'
]);
this.updateGenomesInDisplay([
'Bile', 'BSK30', 'Calcutta', 'Ensete01', 'EnseteBedadit', 'EnseteDerea',
'FHIA', 'Itinerans', 'Kole', 'Lidi', 'MasKirana', 'Pahang', 'PKW',
'Tanduk', 'TongkatLangitMaluku'
]);
this.updateGenomesInDisplaySave([
'Bile', 'BSK30', 'Calcutta', 'Ensete01', 'EnseteBedadit', 'EnseteDerea',
'FHIA', 'Itinerans', 'Kole', 'Lidi', 'MasKirana', 'Pahang', 'PKW',
'Tanduk', 'TongkatLangitMaluku'
]);
//Send datasets to store
let pavDataPromise = d3.json('./bananachePAV_preformatted.json');
let gffDataPromise = d3.json('./bananacheGFF_preformatted.json');
let [pavData, gffData] = await Promise.all( [pavDataPromise, gffDataPromise] );
//Section dedicated to sending the pav updates to the store
this.updateFullChromData(pavData);
this.updateSelectedChrom('chr01');
this.updateFileLoaded(true);
console.log('PAV FILE HAS BEEN LOADED AND CHROMDATA IS SET')
//Section dedicated to sending the gff updates to the store
this.updateFullGffData(gffData);
this.updateIsGffUploadedTRUE();
this.pushSortModeInSortChoice('Gene presence status'); // Add the choice to sort by gene presence status with a search bar
//Section dedicated to the newick phylogeny sent to the store
let newickStr = '((Ensete01,(EnseteDerea,EnseteBedadit)),((TongkatLangitMaluku,(Itinerans,(PKW,(((FHIA,(Bile,Tanduk)),((Kole,MasKirana),(Calcutta,(Lidi,(Pahang,BSK30)))))))))));';
let phyloOrder = this.autoExtractGenoOrderFromNewick(newickStr);
this.updatePhylogenyTree(phyloOrder);
this.updatePhylogenyString(newickStr);
this.pushSortModeInSortChoice('Phylogenetic tree'); // Add the choice to sort by phylogenetic tree
//Disable loadingSpinner in Panache view
this.updateDisplayLoadingStatus();
}Especially, you will want to update the values sent to:
updateChromNames to replace the list of chromosome names,
updateGenomesInDisplay to list the genome names in the same order they appeared within the original pangenome file,
updateGenomesInDisplaySave with the same info to store the original sorting order,
pavDataPromise and gffDataPromise to store your pre-formatted presence absence matrix and gff files, respectively,
updateSelectedChrom to target from this list the chromosome that should be displayed by default,
newickStr to store the newick string used to create the phylogenetic tree for your data.
Once everything is set, you can run your server with one of the two solution detailed earlier, and try your very own version of Panache. Please note that while actively supported, the bananaGenomeHub branch of Panache is not the main one and may be updated less frequently.
Moreover, feel free to raise any concern you may have while using Panache as it can help us improve this documentation for future users.