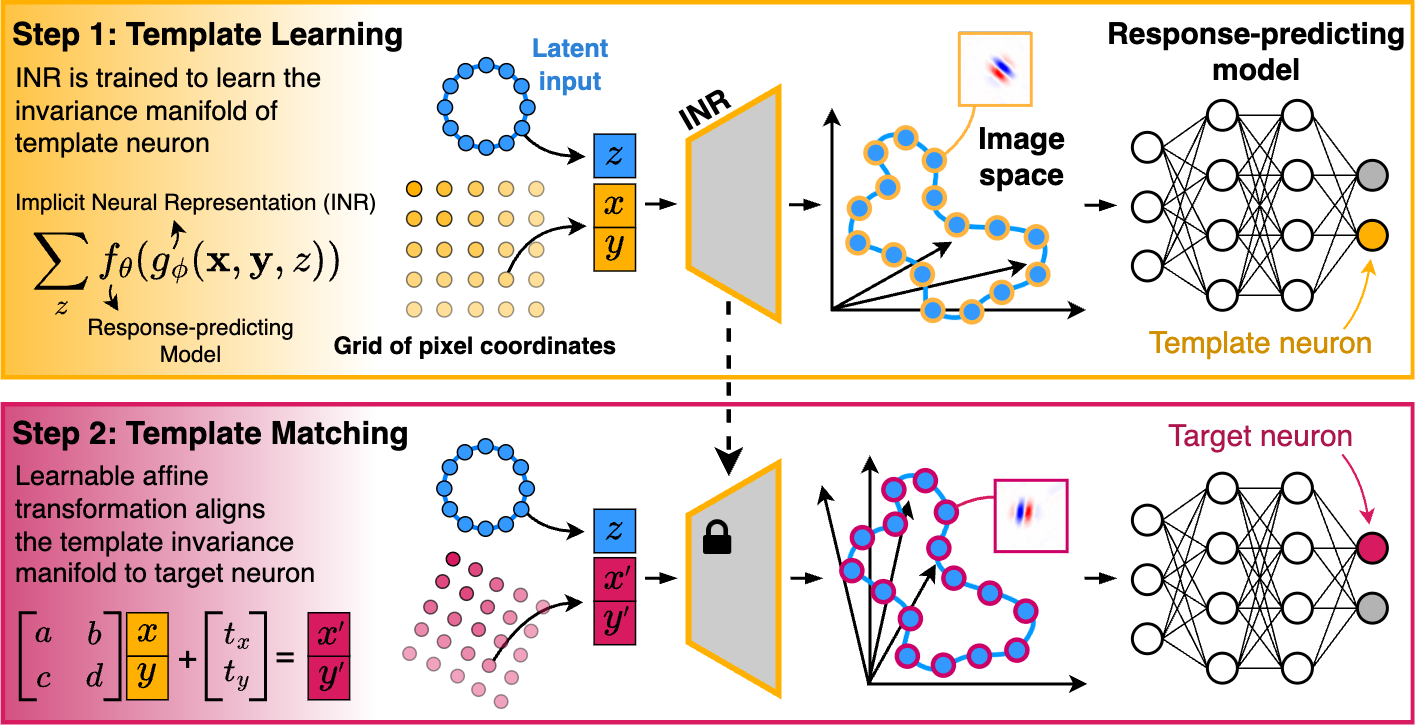
LAMINR (Learning and Aligning Manifolds of Single-Neuron Invariances using Implicit Neural Representations) enables the systematic discovery and alignment of invariance manifolds in stimulus space for visual sensory neurons, providing a principled way to characterize and compare neuronal invariances at the population level, independent of nuisance receptive field properties such as position, size, and orientation.
- Continuous Invariance Manifold Learning: Identifies the full space of stimuli that elicit near-maximal responses from a neuron.
- Alignment Across Neurons: Learns transformations that align invariance manifolds across neurons, revealing shared invariance properties.
- Functional Clustering: Enables clustering neurons into distinct functional types based on their invariance properties.
- Model-Agnostic: Can be applied to any robust response-predicting model of biological neurons.
You can install LAMINR using one of the following methods:
pip install laminrpip install git+https://github.com/sinzlab/laminr.gitHere's a simple example of how to use LAMINR to learn and align invariance manifolds.
from laminr import neuron_models, get_mei_dict, InvarianceManifold
device = "cuda"
input_shape = [1, 100, 100] # (channels, height, width)
# Load the trained neuron model
model = neuron_models.simulated("demo1", img_res=input_shape[1:]).to(device)
# Generate MEIs (Maximally Exciting Inputs)
image_constraints = {
"pixel_value_lower_bound": -1.0,
"pixel_value_upper_bound": 1.0,
"required_img_norm": 1.0,
}
meis_dict = get_mei_dict(model, input_shape, **image_constraints)
# Initialize the invariance manifold pipeline
inv_manifold = InvarianceManifold(model, meis_dict, **image_constraints)
# Learn invariance manifold for neuron 0 (template)
template_idx = 0
template_imgs, template_activations = inv_manifold.learn(template_idx)
# Align the template to neurons 1 and 2
target_idxs = [1, 2]
aligned_imgs, aligned_activations = inv_manifold.match(target_idxs)We have provided a Dockerfile for building an image with LAMINR pre-installed. Ensure that both docker and docker-compose are installed on your system.
Follow the steps below to run LAMINR inside a Docker container with Jupyter Lab.
1. Clone the repository and navigate to the project directory:
git clone https://github.com/sinzlab/laminr.git
cd laminr2. Run the following command inside the directory:
docker compose run -p 10101:8888 examplesThis command:
- Builds the Docker image and creates a container.
- Exposes Jupyter Lab on port 10101.
3. Access Jupyter Lab: Jupyter Lab will launch in the examples folder, which you can open in your browser via localhost:10101 (the token can be found in the terminal logs).
If you encounter any issues while using the method, please create an Issue on GitHub.
We welcome and appreciate contributions to the package! Feel free to open an Issue or submit a Pull Request for new features.
For other questions or project collaboration inquiries, please contact [email protected] or [email protected].
This package is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) License. Briefly:
- Attribution Required: You must credit the original authors and indicate if changes were made.
- NonCommercial Use Only: This package may not be used for commercial purposes without explicit permission.
- No Additional Restrictions: You may not apply legal terms that prevent others from using this package under these terms.
For full details, see the CC BY-NC 4.0 License.
For commercial use inquiries, please contact: [email protected].
ICLR 2025 (Oral): Learning and Aligning Single-Neuron Invariance Manifolds in Visual Cortex
Authors: Mohammad Bashiri*, Luca Baroni*, Ján Antolík, Fabian H. Sinz. (* denotes equal contribution)
Please cite our work if you find it useful:
@inproceedings{bashiri2025laminr,
title={Learning and Aligning Single-Neuron Invariance Manifolds in Visual Cortex},
author={Bashiri, Mohammad and Baroni, Luca and Antolík, Ján and Sinz, Fabian H.},
booktitle={International Conference on Learning Representations (ICLR)},
year={2025}
}-CA3FE6.svg?style=flat)